Why do microbiologists have to invent new terms?!
A recent tweet (see below) brought up a great point and is something I have been thinking about for some time now. With all the new capabilities to assess the HUGE amounts of microbial diversity, I think it is time for some normalization of how we describe and discuss microbes. Just like any systematics in any organism, there are issues. However, the ability to describe evolutionary relationships has been at the foundation of evolutionary biology since Darwin and is integral to how we compare and assess various eco/evo processes affecting biodiversity and biogeography.
At some level, it's easy to differentiate species. For instance, I think everyone can tell the difference between a red algae and a horse. With microorganisms, it isn't as easily to differentiate individuals based on morphological attributes; and instead we lean heavier on genetic and molecular techniques. Commonly, microbiologists use conserved marker genes, such as the 16S rRNA gene, to classify and assess phylogenetic relatedness among isolates and/or culture-indepedent amplicon sequences. This has led to unprecedented progress in our understanding of microbial diversity and the underlying processes at relatively broad genetic resolutions in microbial communities. But just as easily as we can go out and assay thousands of microbial taxa in soil, we can just as easily over-interpret our findings. For instance, there is a desire to prematurely relate microbial studies to the enormous theoretical and empirical work done with macroorganisms. But, microbial ecology is in its infancy and, as such, I find the use of microbe-specific terminology informative and necessary.
OTUs, microdiversity, populations, and species, OH MY!
Now before I start my rant into microbial terminology, I want to point out that macroorganisms have their own confusing terminology (e.g., cryptic species). Ambiguous terminology is always something that will be an issue, but, nevertheless, remains an important topic when analyzing biodiversity patterns. In any case, specifically for microbial ecology and evolution, I think the fundamental issue arises from the inability to relate molecular methods to biological questions. Just as in any study, the molecular methods you choose must reflect your biological question because, as this commentary so nicely puts it, "in nature, there is only diversity" [1]. If we just go out and blindly sample microbial diversity, we cannot address the mechanisms contributing to observed biogeographical patterns.
Microbial species
The fundamental unit of biology is the species. For decades, microbiologists have tried to quantify what constitutes a species and when other clades should be reclassified. For this post, I will only concentrate on the newer techniques used for delineating microbial taxa.
Obviously this metric only examines genomic relatedness and does not consider whether environments or ecology contribute to species designations. I will come back to these ideas later, but for now let's move on to OTUs, or operational taxonomic units.
OTUs or ESVs cannot represent species
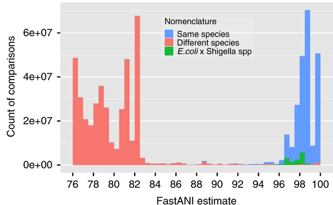
Increasingly, researchers are using culture-independent methods to assess previously unknown microbial diversity. The most popular of these methods is amplicon-based metagenomic analyses utilizing the 16S rRNA marker gene. With newer and newer technologies and computational resources, we can easily sequences hundreds of thousands of microorganisms from a given environmental sample. This has led to the implementation of delineating microbial taxa into operational taxonomic units (OTUs), but it is unclear how these taxonomic delineations represent more traditionally-defined terminology. Initially, some tried to relate OTUs to microbial species but these proved far too broad to quantify. For instance, a 3% divergence in the 16S rRNA gene (the most common threshold for OTU clustering) represents the same evolutionary divergence time as the origin of modern birds or roughly 150M years. Reread that last sentence and let it sink in...
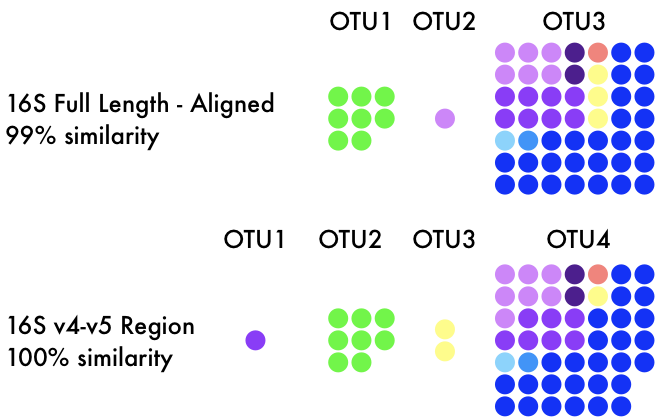
With newer sequencing technologies came the reduction of sequencing breadth of the 16S rRNA gene. Most studies nowadays target a hypervariable region of the 16S rRNA gene (usually the V4/V5 region), which only targets a small section of the ~1600bp 16S gene. This is advantageous as we can ignore conserved, non-informative regions and sequence more individuals within the community. This has also led to a movement away from OTUs to exact sequence variants (ESVs) to quantify microbial diversity. This would allow each variant (or SNP) in the 16S gene to infer evolutionary history, providing a far more realistic depiction of microbial taxonomic units. Now this is where we circle back around again. Repeatedly, I have seen papers inferring "population dynamics" or "intraspecific variation" across microbial taxa by assessing differential distributions of ESVs (sound familiar? remember the birds...). I do not want to list papers, but I fear this is becoming more widely abused in practice as more and more microbial communities are assessed. But just as full genomes do not consider ecological factors, neither ESVs nor OTUs provide the resolution to infer functional traits or how those correspond to environmental factors. And I am being generous, there are examples of a single SNP in the entire genome contributing to diversification. Microdiversity
This is where the term microdiversity can be beneficial. I will define microdiversity as closely-related strains within the same taxonomic group, however defined. For ease of this argument, I will focus on the microdiversity within OTUs or ESVs (hereafter referred to just OTUs since its just a matter of the clustering threshold). As stated above, we know little of the variation within OTUs and how trait variability contributes to the distribution of microbial taxa. Moreover, we do not know how to delineate microbial species and sometimes even genera (see Escherichia and Shigella). Therefore, relating community analyses with corresponding OTU distributions is almost impossible to equate to frameworks devised for macroorganisms at the species level.
Moving towards a bacterial species definition - ecotype model
Incorporating genomic and ecological information (i.e., environmental distributions and phenotypes) allows a more robust picture to emerge for delineating microbial taxa. Cohan proposed adapting an "ecotype model" to delineate ecological populations, as he describes it [5]:
"ecologically distinct lineages (based on sequence analysis) and as a prognosis of future coexistence (based on ecological differences), are the fundamental units of bacterial ecology and evolution"
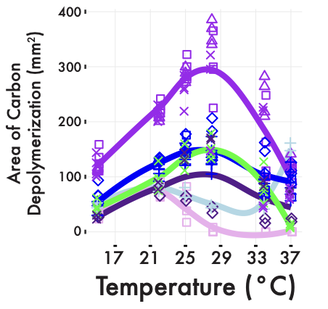
Essentially, he is describing the fundamental unit of biology, species (I know, ecotype means ecological population but I am equating it to species - can it get more confusing!). However, to get to this point requires extensive work in a microbe: collate genomic, phenotypic, environmental data, etc. to delineate ecotypes. And even if you can get to this point, there is still ambiguity to what level qualifies an ecotype as it can be a sliding scale. For instance, the heavily studied marine cyanobacterium, Prochlorococcus has been designated into several ecotypes, each corresponding to phenotypic traits with these traits highly correlated to environmental distributions (see other blog post here). Even in the best-known case of Prochlorococcus, we are still clustering genomes into ecotypes that diverge far below suggested ANI species boundaries (e.g., Pro ecotype HLII includes genomes <88% ANI).
Simply put, there is still A LOT of work to do. Evolutionary Processes and Populations
Lastly, I just want to end with microbial populations. Ideally, this is the level microbiologists need to get if we wish to understand the evolutionary processes driving speciation. Populations, by definition, are groups of individuals within the same species. By "jumping the gun" and moving from environmental community-level studies to "intraspecific variation" using ESVs we are doing ourselves a disservice. We need to learn to walk before we can run. And this requires extensive work to first describe species and then ask questions into the underlying population dynamics driving microbial diversification.
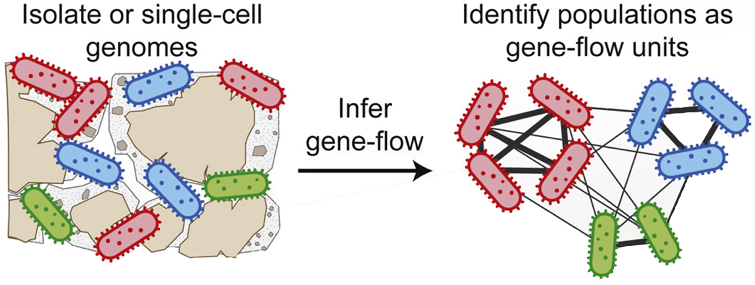
Populations are also groups of interbreeding individuals, but as asexual organisms, microbial are harder to pinpoint. However, we can use recombination as a metric for reproduction to identify possible population clusters. A recent study from MIT attempts to do just that, by defining populations to illuminate ecologically-distinct populations (figure below) [6]. Papers
1. McLaren MR, Callahan BJ. (2018). In nature, there is only diversity. mBio 9: e02149-17.
2. Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. (2019). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nature Communications 9: 5114. 3. Chase AB, Karaoz U, Brodie EL, Gomez-Lunar Z, Martiny AC, Martiny JBHM. (2017). Microdiversity of an abundant terrestrial bacterium encompasses extensive variation in ecologically relevant traits. mBio 8: e01809-17. 4. Chase AB, Gomez-Lunar Z, Lopez AE, Li J, Allison SD, Martiny AC, Martiny JBHM. (2018). Emergence of soil bacterial ecotypes along a climate gradient. Environmental Microbiology 20: 4112–4126. 5. Cohan FM. (2006). Towards a conceptual and operational union of bacterial systematics, ecology, and evolution. Philos Trans R Soc Lond B Biol Sci. 361: 1985-1996. 6. Arevalo P, VanInsberghe D, Elsherbini J, Gore J, Polz MF. (2019). A reverse ecology approach based on a biological definition of microbial populations. Cell 178: 820-834.e14.
0 Comments
Leave a Reply. |
AuthorSome thoughts on some (small) things Archives
May 2023
Categories |
Proudly powered by Weebly
 RSS Feed
RSS Feed