Dr. Alexander Bogdanov, and Dr. Chase deploying resin bags in Cabrillo National Monument, San Diego, CA. Credit: Devin Oktar Yalkin, Scientific American.
June 2024 New paper in Nature Comm discovering new natural products from intertidal regions
New natural products (June 2024)Two new papers on marine natural products. The first, led by Dr. Doug Sweeney, investigates the potential for a marine lineage of Streptomyces (MAR4) to be a treasure trove of new natural products (the first of many paper to come from these amazing bacteria, stay tuned!). The second, led by Dr. Alexander Bogdanov, was a monumental and collaborative effort consisting of our fieldwork at Cabrillo (see more below) to discover new carbon skeletons using a non-invasive sampling technique in intertidal, near-shore regions.
|
Presenting data!! (April 2024)Chase Lab members were busy presenting data at a range of conferences and symposia. Our undergraduate, Ruhani (pictured) presented some new work investigating the potential of methanogens to remediate natural gas pipeline leaks. Prof. Chase presented some work on the evolution of microbiomes at EGU in Vienna, Austria as part of a special session entitled, "Eco-Omics: Harnessing meta-omics to understand the biogeosciences across scales: from the cell to Earth system". Also gave an invited seminar at OSU Micro.
|
New collaborations at SMU (July 2023) |
Chemical ecology (April 2023) |
|
Our new work with Dr. Kathleen Smits (SMU Engineering) is a multi-disciplinary approach to understand carbon sinks and sources in soils. Notably, we are working on methane mitigation from anthropogenic subsurface sources, including natural gas pipeline leaks, that are dispersed and variable. By combining field experiments and collaborative efforts, we hope to leverage methane contaminant fate and transport with the metabolic diversity of soil methanotrophs. Initial fieldwork began this summer at CSU's METEC.
|
New paper out in The ISME Journal investigating the role of biotic interactions on microbiome community structure. While it's thought that biotic interactions (e.g., competition, predation) drive community assembly at local scales, this is difficult to test in microbial communities. One way we circumvented this process was to look at the production of specialized metabolites and the distributions of their biosynthetic machinery. For more info, check out our editorial featured in Nature Microbiology.
|
Evolution in biogeochem (March 2023) |
New position at SMU (April 2022) |
|
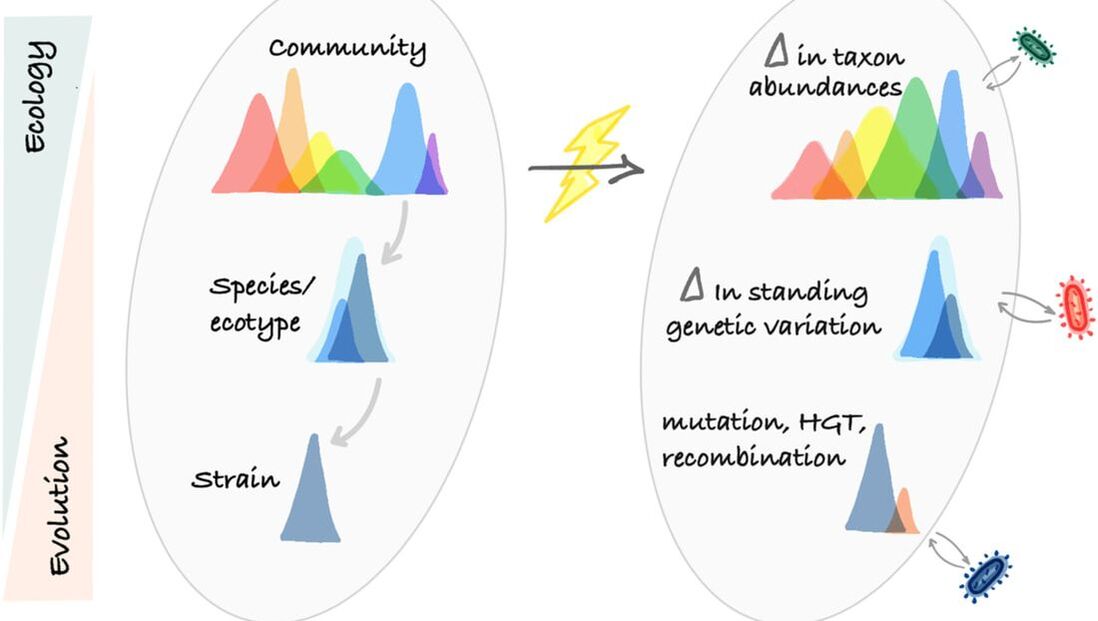
Two new papers on the importance of quantifying microbial evolution in response to environmental change. In the first paper, published in EMI, we addressed knowledge gaps to advance current modeling approaches with respect to microbial eco-evolutionary mechanisms in soil C models. In our second paper, published in Ecology Letters, we proposed a conceptual framework for explicitly integrating evolution into microbiome–functioning relationships.
|
Excited to announce I will be joining the Department of Earth Sciences at SMU in the Fall of 2022. This will be part of a cluster hire to incorporate climate and atmospheric science to address anthropogenic induced climate change. My lab will be focusing on microbial ecology and evolution in the context of biogeochemical cycling in environmental systems. I am looking for motivated people to join the lab, so feel free to contact me for more information!
|
First NP paper in mBio (Nov 2021) |
NIH R21 Fieldwork (August 2021) |
|
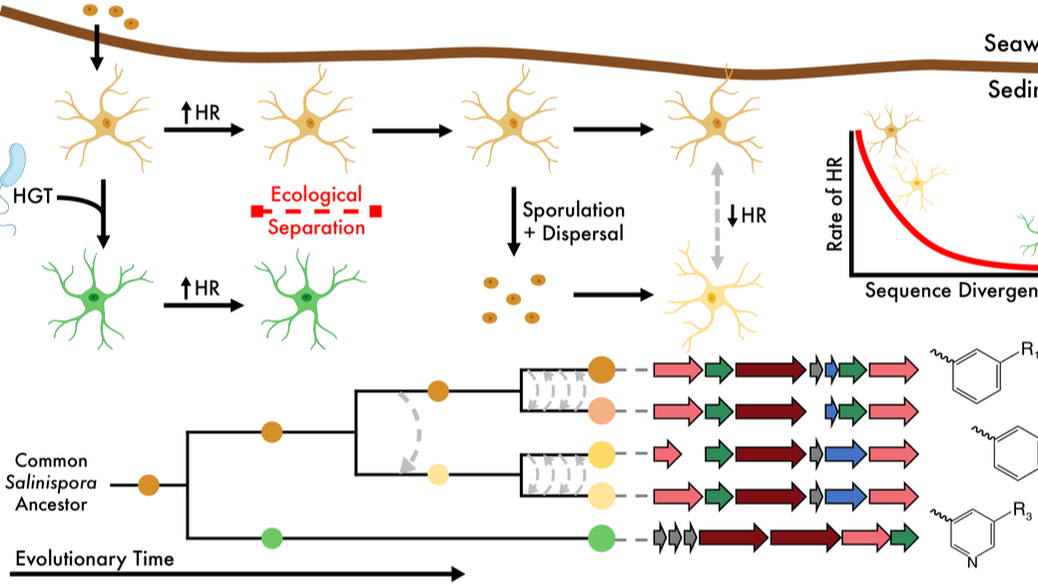
Coming from a background in eco/evo, I found it interesting how natural products paper focused so much on horizontal gene transfer of entire BGCs (that can be very large!) across disparate taxa. This paper is the result of looking at how specialized metabolites, and the cognate BGCs, evolve once acquired. Spoiler alert: BGCs are retained over evolutionary time through processes of vertical inheritance. Through this retention, evolutionary processes can act on the biosynthetic genes to generate new chemical diversity.
|
We started collecting intertidal samples at Cabrillo National Monument in San Diego. Working with the National Park Service has been awesome (featuring some of the best views), plus I got to reunite with some Anteater friends, Drs. Pandori and LA Cat. We are looking for novel marine natural products using a resin capture technique. Pictured is postdoc, Dr. Alex Bogdanov, from the Jensen Lab deploying our samples with downtown SD in the background. Our preliminary data looks like we have some new halogenated molecules!
|
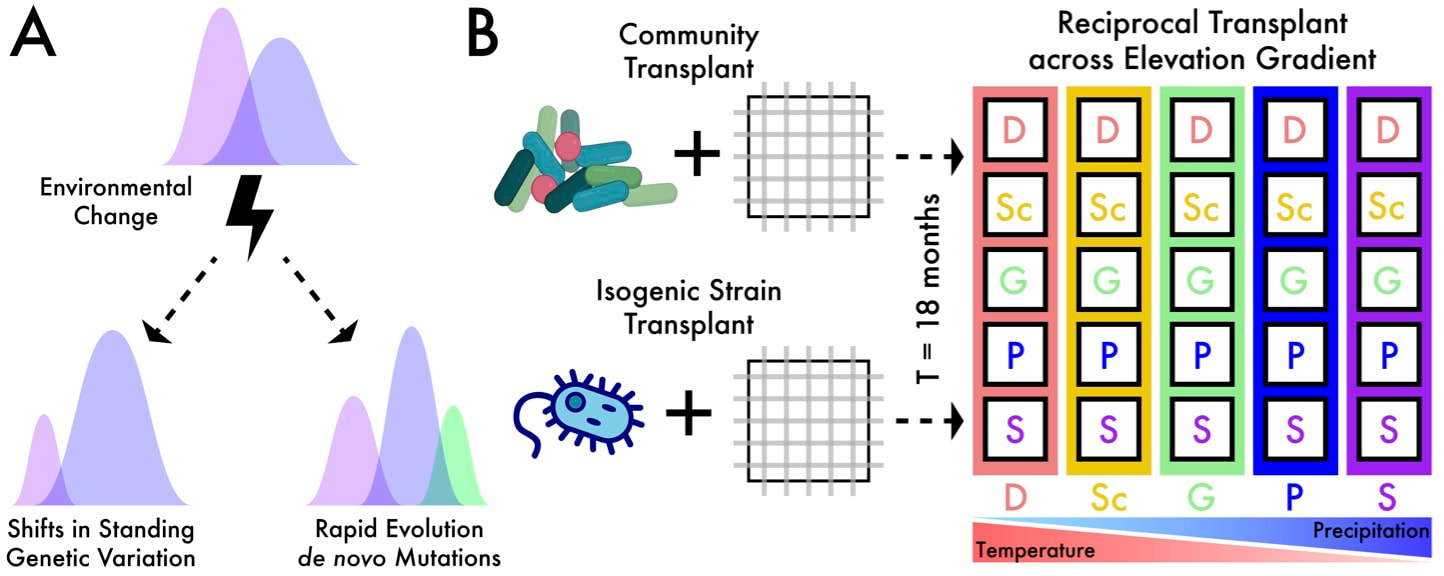
New paper in PNAS! (May 2021)Our recent publication about bacterial evolution was published in PNAS. This was a HUGE effort by everyone in the Martiny Lab (at UCI) to conduct this large-scale reciprocal transplant experiment across a regional climate gradient. We concentrated on the responses of bacterial "species" but you can also check out the community-level analyses here.
See the press release and news coverage as well. |
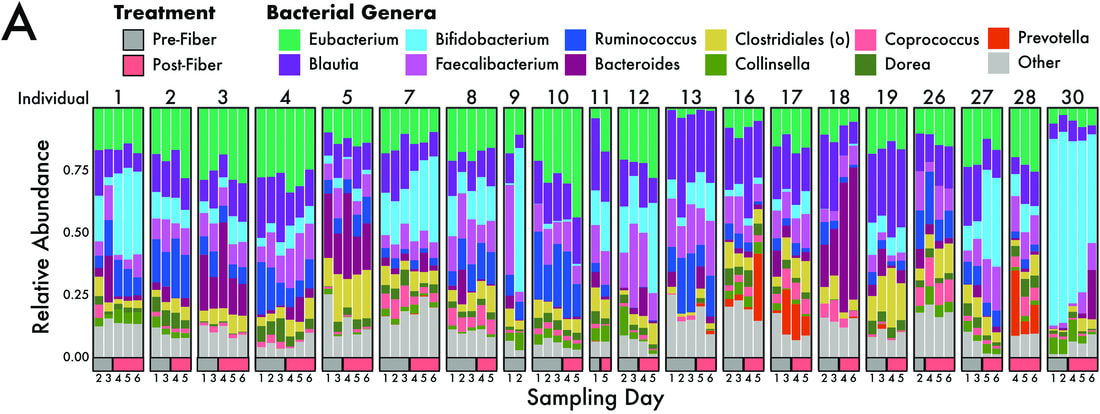
May the fiber be with you (March 2021)We published the results from our CURE course where undergrads voluntarily underwent a fiber intervention in their diets by increasing to 50g a day! We saw compositional shifts in the gut microbiome associated with dietary shifts, including an increase in fiber degrading bacteria. Want to change your gut microbiome? It's possible in only two weeks!
|
Bacteria and sympatry (October 2019)While we are still trying to figure out what constitutes bacterial species and populations, we published a new paper in mBio identifying the factors structuring the genetic diversity of a soil bacterium across varying spatial scales. We found that bacterial populations are maintained both by ecological specialization within localized microenvironments and by dispersal limitation between geographic locations.
|
New fellowship at SIO (January 2019)I am starting a postdoc with Paul Jensen at SIO to integrate evolutionary biology into the discovery of marine natural products. My fellowship was awarded by the Scripps Postdoctoral Scholar Program for two-years and I am super excited to get started! I will be applying a trait-based framework to identify the processes structuring the diversity and distribution of specialized metabolites.
|
Proudly powered by Weebly