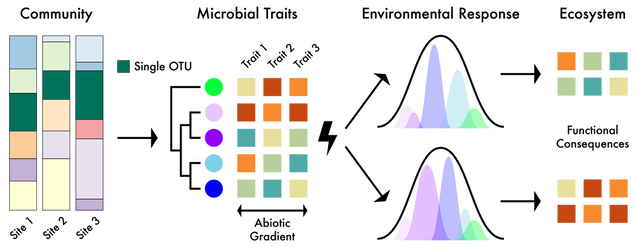Overview
What are the processes contributing to the maintenance of microbial diversity?
This is the main question our research aims to address. To do so, we have spent a lot of time thinking about the appropriate biological resolution to accurately assess and quantify these processes in microbiomes (Martiny et al. 2023). By establishing the correct biological unit (i.e., populations and species) that are used in macro-organisms, we assess the ecological and evolutionary processes driving microbial diversity and distribution. In the past, our work has largely focused on the abiotic, environmental factors contributing to the observed geographic distributions of microbes by correlating functional trait variation to resource and niche partitioning. Recently, we shifted to the traits mediating biotic interactions between microbes, focusing on the production of natural products. These specialized metabolites, which include antibiotics and siderophores, can aid in direct competition, nutrient uptake, and defense and likely represent an important, yet understudied, collection of functional traits. Ultimately, our works aims to identify the underlying functional traits within microbial taxa that contribute to their distribution, and understand whether this variation (or the processes driving this variation) is relevant at the ecosystem level.
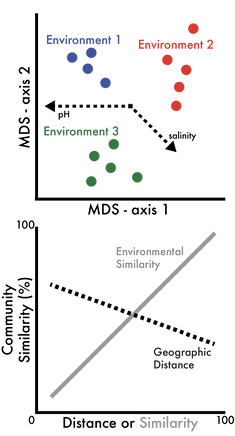
Biogeography allows for insights into the eco/evo processes influencing the spatial distribution of biodiversity and the environmental factors that can contribute to community assembly. Traditionally, these patterns in macroorganisms (e.g., plants and animals) describe the spatiotemporal distributions at the species or population level. Conversely, microbial studies often concentrate at the community level due to the ease of sequencing entire microbiomes. But by doing so, these broad community characterizations collapse microbial genetic diversity into operational taxonomic units (OTUs), thereby limiting the detection of finer-scale genetic and phenotypic variation that contributes to biogeographic distributions (Chase and Martiny, 2018). However, it is at this finer level of genetic differentiation where we can correlate microbiome response to environmental change to better understand the influence on ecosystem level functions, such as soil carbon cycling (Abs et al. 2023).
|
Ecological Responses. It is now well established that various biotic and abiotic factors influence microbial biogeographic patterns at the community level. However, the mechanisms maintaining these patterns are largely unknown. We are interested in linking the fine-scale genetic variation among closely-related taxa to variation in ecologically-relevant traits (Chase et al. 2017). Variation in these traits should translate to differential distributions across environments, where the environment selects for specific clades (and traits) under variable conditions. By delineating ecotypes, or fine-scale genetic clusters sharing ecologically-relevant phenotypic traits, we can map environmental distributions to link trait variation to spatial distributions (Chase et al. 2018). Inevitably, we are interested in elucidating the key environmental parameters contributing to niche differentiation.
We are also interested in processes that are independent of environmental selection, including stochastic or neutral processes, that also contribute to community dynamics. By controlling dispersal with field manipulation experiments, we can quantify the effects of stochasticity on the beta-diversity in bacterial communities (Albright et al. 2019). Our observations indicated that stochastic effects on beta-diversity were not attenuated at the functional level, as measured by genetic functional potential and extracellular enzyme activity. |
Evolutionary Responses. While we can readily observe the ecological responses by microbial communities, it remains unclear how evolutionary responses influence shifts in microbial diversity (Martiny et al. 2023). Microbes, with their short generation times and large population sizes, may respond quickly to new environments through adaptive evolution. Certainly, laboratory studies have repeatedly demonstrated rapid evolutionary adaptation in bacterial populations with consequences for organismal physiology. Yet, it remains unclear how in vitro observations of evolutionary processes extend to in situ communities and whether the rapid adaptive responses observed in laboratory studies is relevant on ecological timescales. By using a large-scale reciprocal transplant experiment, we demonstrated the potential for rapid evolutionary change (within 18 months) by a soil bacterium in response to natural environmental variation on the same timescale as typically observed for ecological processes (Chase et al. 2021). Our field experiments demonstrate how both demographic shifts of ecotypes and contemporary evolution can alter the diversity and functioning (C degradation) of a soil microbiome on the same timescale (Abs et al. 2024).
Population dynamics, including shifts in allele frequencies, can also provide fitness benefits that gradually impact evolutionary trajectories. And while de novo mutations can lead to novel adaptations, gene flow within and between populations can also contribute to local environmental adaptation and diversification. Using comparative genomics, we are interested in how phylogenetic structure within a single bacterial ecotype can be used to infer patterns of gene flow (Chase et al. 2019). Our results indicate that the genetic structure among populations are maintained both by ecological specialization in localized microenvironments (isolation by environment) and by dispersal limitation between geographic locations (isolation by distance).
Population dynamics, including shifts in allele frequencies, can also provide fitness benefits that gradually impact evolutionary trajectories. And while de novo mutations can lead to novel adaptations, gene flow within and between populations can also contribute to local environmental adaptation and diversification. Using comparative genomics, we are interested in how phylogenetic structure within a single bacterial ecotype can be used to infer patterns of gene flow (Chase et al. 2019). Our results indicate that the genetic structure among populations are maintained both by ecological specialization in localized microenvironments (isolation by environment) and by dispersal limitation between geographic locations (isolation by distance).
Natural Products and Biosynthetic Gene Cluster Evolution
While environmental (i.e. abiotic) factors govern observed distributions of the taxa within microbial communities, less is known about the biotic interactions occurring between microbes. Likely, these species interactions are largely mediated through a chemical network within these complex and highly diverse microbial communities. However, we know little of the ecological aspects of secondary metabolites or how the molecules evolve and contribute to spatial distributions. Viewing the production of microbial natural products as functional traits driving differentiation among bacterial lineages facilitates an ecological perspective into the distribution of potentially new bioactive molecules. For instance, our recent work has shown that environmental microbiomes contain unique assemblages of bioactive enzymes in a habitat-specific manner (Singh et al. 2023).
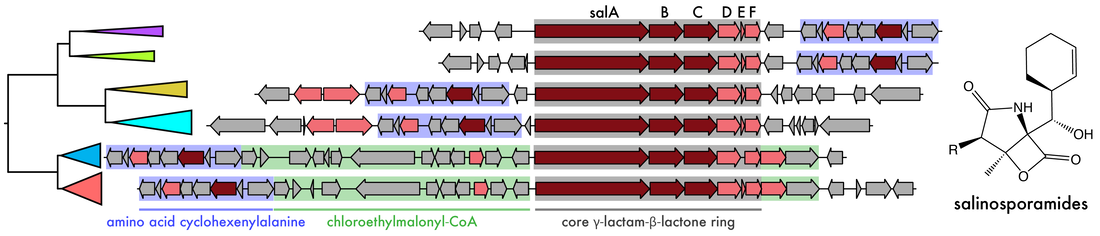
By investigating the prolific natural product producing bacterium Salinispora (Román-Ponce et al. 2020), we established that biosynthetic potential and the production of specialized metabolites can be viewed as functional traits driving species diversification. While previous evidence indicated that horizontal gene transfer (HGT) largely contributed to BGC diversity, we found that a majority of BGCs in Salinispora genomes are maintained by processes of vertical descent (Chase et al. 2021). Applying targeted high-resolution tandem mass spectrometry (LC-MS/MS) of experimentally characterized BGCs, we identified a range of evolutionary processes facilitated by vertical inheritance, including gene gain/loss events and constrained intraspecies recombination, as contributors to BGC diversification. Notably, the evolutionary processes driving BGC diversification had direct consequences for compound production, providing insights into the evolutionary processes generating new chemical diversity. Resolving these evolutionary relationships between closely related strains and specialized metabolism will facilitate our understanding of the ecological roles of these small molecules and their influence on community structure.
|
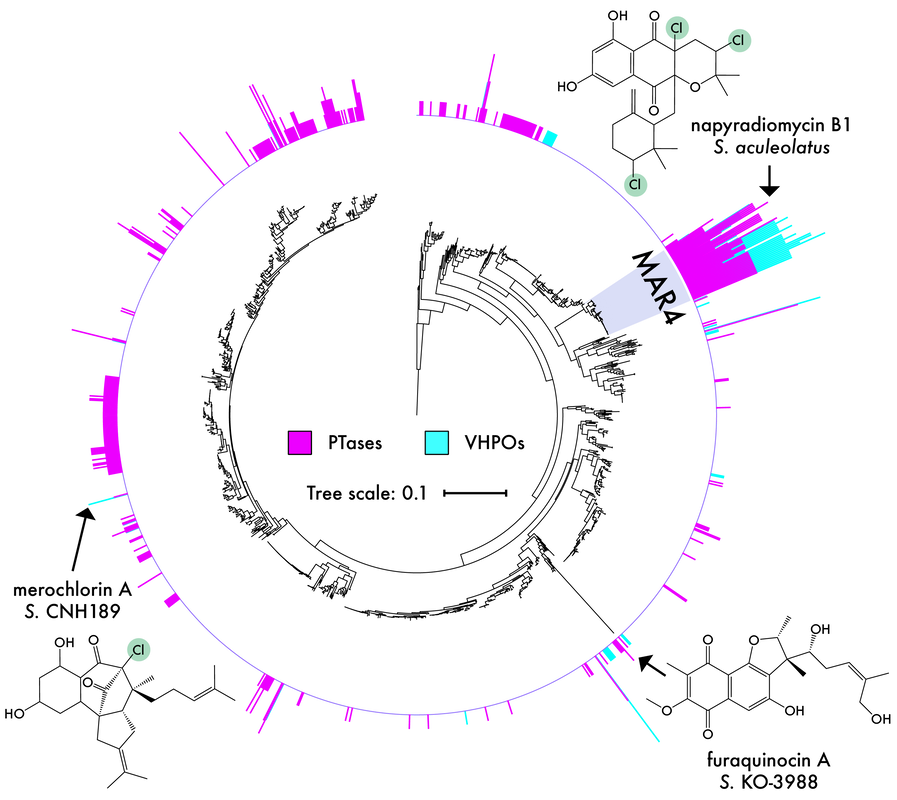
By expanding a trait-based approach to other bioactive taxa, we can effectively target biochemically novel lineages for focused natural product discovery efforts. Similar to work in soils, marine sediment actinomycetes, particularly Streptomyces, have become the most prolific source of antibiotics identified to date. Despite the deep history and intensive work done in Streptomyces, we have identified a novel marine lineage, MAR4, that are enriched in genes that encode the biosynthesis of hybrid isoprenoids (i.e., prenyltransferases; PTases) and vanadium-dependent chloroperoxidase (VHPO) enzymes (Sweeney et al. 2024). Most of the MAR4 natural products are known for their antibacterial properties, such as the napyradiomycin family of meroterpenoids that exhibit potent and rapid bactericidal activity. However, the biosynthesis of many of the known molecules, particularly in relation to the BGCs that encode them, remains ambiguous. We are currently working to link the observed genomic signatures in MAR4 strains to their chemical diversity, with a focus on halogenated compounds.
|
Establishing that the production of specialized metabolites represent a conserved phylogenetic trait differentiating bacterial lineages allows for the incorporation of biogeography to discover new pharmaceutical drug leads by scaling from individual taxa to entire microbial communities. For instance, at large spatial scales, we find that community differences reflect variable biosynthetic enzymes (Singh et al. 2023); whereas at smaller spatial scales, ecological theory predicts that biotic filters, such as predation, competition, and species interactions, will select taxa from a regional species pool to comprise the local community (see our commentary here). Through the use of environmental metabolomics, we can characterize the complex metabolome to demonstrate that fine-scale differences among community members reflects extensive variation in the composition of biosynthetic enzymes and gene clusters, metagenome-assembled genomes (MAGs) enriched in these BGCs, and the production of metabolites (Chase et al. 2023). In addition, this paired-omic environmental approach promotes the discovery of novel metabolites from these environments in an organism-independent manner (Bogdanov et al. 2023).
Microbes reside in communities that, on the aggregate, form microbiomes. Whether these are environmental microbiomes or human-associated, both biotic and abiotic factors regulate community assembly and composition. By viewing microbiomes through the lens of community ecology, we can also investigate the processes impacting the composition of the human microbiome. For example, we identified pH as an important environmental factor in lung microenvironments for colonization and adaptation of the cystic fibrosis pathogen, Stenotrophomonas maltophilia (Gallagher et al. 2019).
We can also apply this framework to ask all sorts of human health questions like: how does diet alter the human gut microbiome. Notably, we approached this topic from a unique perspective by integrating contemporary research with education. Specifically, we established a course-based undergraduate research experience (CURE) at UC Irvine in collaboration with the UCI Microbiome Initiative to assess the effects of a fiber intervention on the gut microbiomes of undergraduate students (Sewall et al. 2020). Outside of the educational benefits for students, we also observed the shift in diet resulted in significant changes to the composition of individual gut microbiomes (Oliver et al. 2021). Notably, microbial taxa that increased in relative abundance as a result of the diet change included known microbiota-accessible carbohydrates (MACs) degraders (i.e., Bifidobacterium and Lactobacillus). However, these taxonomic shifts did not correspond to consistent shifts in the breakdown of fiber to short-chain fatty acids, as detected by targeted gas chromatography-mass spectrometry (GC-MS).
We can also apply this framework to ask all sorts of human health questions like: how does diet alter the human gut microbiome. Notably, we approached this topic from a unique perspective by integrating contemporary research with education. Specifically, we established a course-based undergraduate research experience (CURE) at UC Irvine in collaboration with the UCI Microbiome Initiative to assess the effects of a fiber intervention on the gut microbiomes of undergraduate students (Sewall et al. 2020). Outside of the educational benefits for students, we also observed the shift in diet resulted in significant changes to the composition of individual gut microbiomes (Oliver et al. 2021). Notably, microbial taxa that increased in relative abundance as a result of the diet change included known microbiota-accessible carbohydrates (MACs) degraders (i.e., Bifidobacterium and Lactobacillus). However, these taxonomic shifts did not correspond to consistent shifts in the breakdown of fiber to short-chain fatty acids, as detected by targeted gas chromatography-mass spectrometry (GC-MS).
Collaborators
|
Jennifer Martiny - UC Irvine [link]
Paul Jensen - Scripps Institution of Oceanography [link] Steve Allison - UC Irvine [link] Eoin Brodie - Lawrence Berkeley National Laboratory [link] Ulas Karaoz - Lawrence Berkeley National Laboratory [link] Adam Martiny - UC Irvine [link] Bradley Moore - Scripps Institution of Oceanography [link] Martin Polz - University of Vienna, formerly MIT [link] Alejandra Rodriguez-Verdugo - UC Irvine [link] Kathleen Smits - Southern Methodist University [link] Katrine Whiteson - UC Irvine [link] |
|
Proudly powered by Weebly