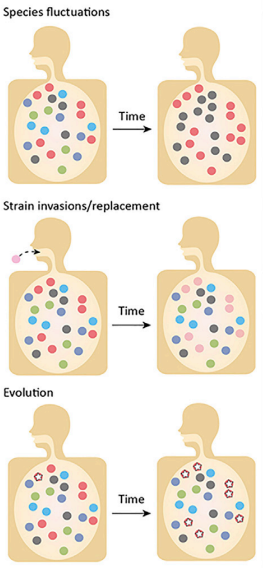
Introduction to Evolutionary processesThe microbiome is the aggregation of all the microorganisms in a system; this can be the human gut microbiome or a biofilm on a rock in a stream. Regardless of the system, the microbes inhabiting that environment are constantly competing for resources (e.g., physical space, nutrients, etc.). Thus, community assembly can be highly deterministic if the environmental conditions and interspecific interactions determine which species persist in a local community. Typically, we associate microbe's traits to the environment, as these traits underlie an organism's response to both biotic and abiotic factors. These traits are inherently based on evolutionary history. So a major question is how can we observe evolution in these systems? For one, environmental disturbances can lead to community turnover - this is typically associated with ecological processes. However, is it time to start reevaluating these processes from an evolutionary standpoint? After all, a change in allele frequency is the definition of evolution. Typically, evolution is studied at the population level, but this is increasingly hard to do in microbiomes. What defines a natural population? Moreover, with rapid population sizes and fast generation times, how can we disseminate ecological and evolutionary processes in microbiomes? Maybe they are one in the same...This month's reading list will highlight some recent (and not so recent) papers addressing evolutionary processes and how they relate to microbiomes. I will not go into population-level processes since I have covered them in other posts here and here. Instead, I want to focus on the challenges in microbiome studies on the aggregate. Human Microbiome
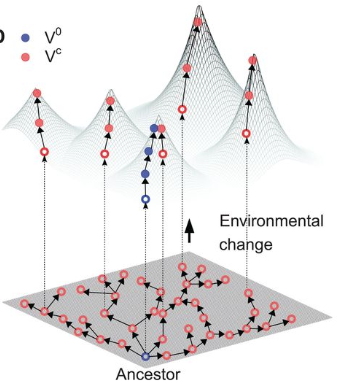
Niche expansion and adaptationIf mutations can lead to increased fitness, what prevents the "optimal" phenotype from dominating a given system. Certainly, a generalist approach can utilize diverse resources and outcompete specialists with limited resources and narrower niche breadth. The next paper address the idea of evolvability [2] with respect to the generalist v. specialist strategy. In particular, the cost of a generalist resides in their reduced capacity to evolve to changing environments. In other words, selection is weakened or relaxed in the generalist strategy since it can readily access a wider range of niches, albeit at local peaks of lower fitness. The authors describe this in the context of a fitness landscape where specialists can rapidly respond to environmental changes whereas the generalist must maneuver through a rugged fitness landscape resulting in a "lag load" to peak fitness.
Observing evolution and molecular adaptationAll of this has been amazing work and definitely worth pursuing more in microbial systems. But a major question is how to observe these evolutionary dynamics in high resolution. This last paper utilizes a barcoding system to track adaptation in yeast [5]. Using this method, the authors can re-barcode each population to tract specific lineages arising from mutations. Initially the ancestral population accumulates a distribution of beneficial mutations. Lower fitness clones die out and are replaced by newer, other beneficial mutants that maintain diversity in the population. This distribution of continuously generated variation provides a broad distribution across a fitness mean. Papers:1. Garud NR, Pollard KS. (2019). Population genetics in the human microbiome. Trends in Genetics
2. Bono LM, Draghi JA, Turner PE. (2019). Evolvability costs of niche expansion. Trends in Genetics 3. Venkataram S, Monasky R, Sikaroodi SH, Kryazhimskiy S, Kacar B. (2019). Evolutionary stalling in the optimization of the translation machinery. bioRxiv 4. Zheng J, Payne JL, Wagner A. (2019). Cryptic genetic variation accelerates evolution by opening access to diverse adaptive peaks. Science 365: 347-353. 5. Nguyen Ba AN, Cvijovic I, Rojas Echenique JI, Lawrence KR, Rego-Costa A, Liu X, Levy SF, Desai MM. (2019). High-resolution lineage tracking reveals travelling wave of adaptation in laboratory yeast. Nature 575: 494–499.
0 Comments
|
AuthorSome thoughts on some (small) things Archives
May 2023
Categories |
Proudly powered by Weebly
 RSS Feed
RSS Feed