|
Hey everyone. I know it has been a long time since my last post, but I got completely consumed with work and adjusting to the new "norm" as everyone else has. I know this is not a unique situation for me, but definitely takes a lot out of someone. I am excited to resume some blogging and highlighting some recent papers I come across. I will try and keep these regularly updated, and if anyone has any requests or recommendations, please send them my way - it is much appreciated! In any case, one project, in particular, that has taken up a large portion of my time is looking at how specialized metabolism contributes to bacterial differentiation. From finishing my PhD a couple years ago, I wanted to integrate biotic interactions into microbial biogeography. Most of my work so far has focused on the abiotic factors contributing to environmental distributions. However, these are pretty broad measurements and observed across pretty distinct ecosystems. As a side note, we have some pretty exciting results coming soon describing evolutionary adaptation to abiotic factors as well for Curtobacterium in soils. Getting back to it. These biotic interactions are mediated through the production and secretion of specialized metabolites. These metabolite-mediated interactions are pretty well-documented in plants and animals, but they are pretty difficult to discern for microbes. Rather, these specialized metabolites have been almost exclusively viewed as sources for new natural products. While they have been instrumental for human health applications (i.e., pharmaceutical leads), microbes produce a wide variety of molecules with distinct biological properties mediating antagonistic interactions (i.e., antibiotics), resource acquisition (siderophores), or mutualistic interactions. As such, their abundance in nature and their implied ecological importance suggests these molecules should contribute to differentiation. My recent work is bridging the work from the natural product realm to integrate ecology and evolutionary theory as they relate to microbial diversification. And while I hope my work will be available to read soon (fingers crossed), I wanted to summarize some of the key papers and/or inspirational papers that contributed to my project. From biosynthetic genes to molecular innovation
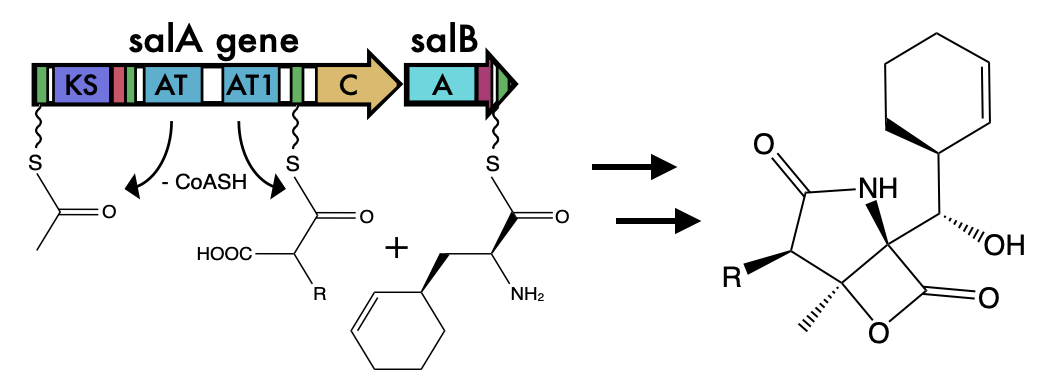
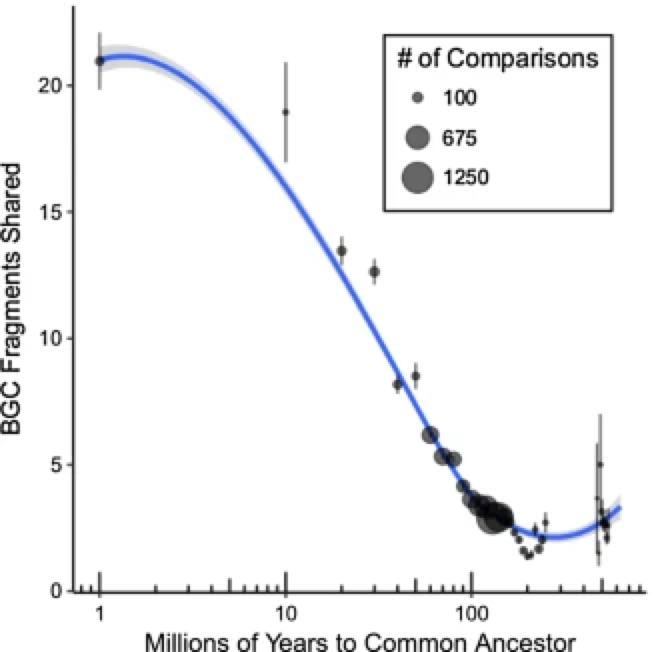
Examination of BGCs can reveal unique enzymology or biochemical interactions to prioritize BGCs for drug discovery. However, what is often ignored in the field of natural products, is how these BGCs evolve. This is of upmost importance because the evolutionary processes governing BGC diversity and distribution should contribute to molecular diversification and, possibly, affect their bioactivity. Within the last 10 years these questions have started to be asked in the field and found that horizontal gene transfer was a major contributor to BGC evolution. Surprisingly, these gene clusters, which can be quite large 50-90kbp, are thought to rapidly be gained and lost through HGT. The repeated observations for evidence of BGCs supports the ideas that these BGCs, and their cognate molecules, are integral to local adaptation. Here is a quote from a paper in 2016 (1): As a collective, the microbial genomes in each environment provide raw materials, the genomic building blocks that individuals can mix and match to create the BGCs that provide selective advantages. Adding ecological considerations to genetic and molecular studies allows the selection principles to be identified and fills out a molecule’s natural history, providing mechanistic insight into the manner in which BGCs are acquired and evolve. These reports were further supported in a systematic review of BGCs that concluded they have been "transferred horizontally at high frequency" (2). Certainly, BGCs can be acquired and result in drastic changes to an organisms' ecology and potentially allow for species transcending events. However, I was always intrigued by the extraordinary capability for these events to occur. The transfer of large, contiguous gene clusters across divergent organisms seems improbable, especially when the costs of HGT are considered ranging from integration of foreign DNA to the regulatory expression to the inherent risk of producing a toxic molecule (for more on HGT costs, definitely read this paper (3)). Despite all of this, the idea that HGT was a major contributor to BGC evolution persisted. After all, these BGCs must have come from somewhere, hence, they must have been acquired at some point in a lineage's evolutionary history. The identification of genomic islands in my third favorite Actino (everyone can probably guess who is #1), Salinispora, further provided a means for this mechanism (4). BGCs can evolve in a "plug and play" model for evolution to confer selective fitness advantages and be highly dependent on the local environment (5). Alternative models for BGC evolutionAt the same time, at least in Salinispora, there is evidence that the molecules are linked to species differentiation; for example, certain molecules are produced exclusively by a single species. This is akin to chemotaxonomy that is widely used in plants, and could also be a contributing factor in bacteria as well. This brings up quite the story. How can BGCs be frequently exchanged and we still observe species-specific signatures in molecule production. One way to look at this is to examine the distribution of BGCs among closely-related strains (stay tuned!!!). If HGT was THE major driver for BGC evolution, we would expect the distribution of BGCs to be random, that is strains that are more phylogenetically similar would have almost no correlation to BGC diversity. However, more and more recent evidence is coming out describing these BGCs (6) as conserved. So how can we reconcile these two seemingly contrasting models for BGC evolution? This is where the recent literature is super exciting, mainly a couple reviews from MG Chevrette (6,7). And the following quote summarizes some of these ideas: Rampant LGT of BGCs is a prevailing hypothesis in the field, but this view often overlooks the large evolutionary timescales in which an LGT event is observed. Vertical inheritance of BGCs represents an alternative mechanism. This is where I think the key is to resolving these ideas and can broken down into two main arguments: 1. HGT is not well-defined and vertical inheritance governs most of BGC diversity 2. BGCs can be readily modified through evolutionary processes to create diverse products
Second, BGCs can evolve de novo. Recent reviews have discussed these ideas and put forward that BGCs evolve neutrally, existing in generalist states to produce a diverse set of products (8). Once BGCs have evolved bioactivity, there are "endless modifications" of the ancestral pathways to increase potency and adapt to environmental conditions (9). So what are these modifications and what processes are governing BGC evolution? A lot of my ideas are presented in my upcoming manuscript that attempts to reconcile these observations to integrate evolutionary theory into natural products research. Briefly, rare HGT events occur and can contribute to large transcendent events. However, the vast majority of processes structuring BGC diversity and distribution are dictated by processes of vertical descent. At this level, fine-scale evolutionary processes (i.e., selection and recombination) can further drive BGC diversification with consequences for molecular innovation. Hope you enjoyed the read! Papers:1. Ruzzini AC & Clardy J. (2016). Gene flow and molecular innovation in bacteria. Current Biology.
DOI: 10.1016/j.cub.2016.08.004 2. Medema MH, Cimermancic P, Sali A, Takano E, Fischbach MA. (2014). A systematic computational analysis of biosynthetic gene cluster evolution: lessons for engineering biosynthesis. PLoS Computational Biology. DOI: 10.1371/journal.pcbi.1004016 3. Baltrus DA. (2013). Exploring the costs of horizontal gene transfer. Trends in Ecology and Evolution. DOI: 10.1016/j.tree.2013.04.002 4. Ziemert N, Lechner A, Wietz M, Millán-Aguiñaga N, Chavarria KL, Jensen PR. (2014). Diversity and evolution of secondary metabolism in the marine actinomycete genus Salinispora. PNAS. DOI: 10.1073/pnas.1324161111 5. Letzel AC, Li J, Amos GCA, Millán‐Aguiñaga N, Ginigini J, Abdelmohsen UR, Gaudêncio SP, Ziemert N, Moore BS, Jensen PR. (2017). Genomic insights into specialized metabolism in the marine actinomycete Salinispora. Environmental Microbiology. DOI: 10.1111/1462-2920.13867 6. Chevrette MG, & Currie CR. (2019). Emerging evolutionary paradigms in antibiotic discovery. Journal of Industrial Microbiology & Biotechnology. DOI: 10.1007/s10295-018-2085-6 7. Chevrette MG, Gutiérrez-García K, Selem-Mojica N, Aguilar-Martínez C, Yañez-Olvera A, Ramos-Aboites HE, Hoskisson PA, Barona-Gómez F. (2020). Evolutionary dynamics of natural product biosynthesis in bacteria. Natural Products Reports. DOI: 10.1039/C9NP00048H 8. Noda-Garcia L & Tawfik DS. (2020). Enzyme evolution in natural products biosynthesis: target- or diversity-oriented? Current Opinion in Chemical Biology. DOI: 10.1016/j.cbpa.2020.05.011 9. Fewer DP & Metsä‐Ketelä M. (2020). A pharmaceutical model for the molecular evolution of microbial natural products. The FEBS Journal. DOI: 10.1111/febs.15129
0 Comments
|
AuthorSome thoughts on some (small) things Archives
May 2023
Categories |
Proudly powered by Weebly
 RSS Feed
RSS Feed