|
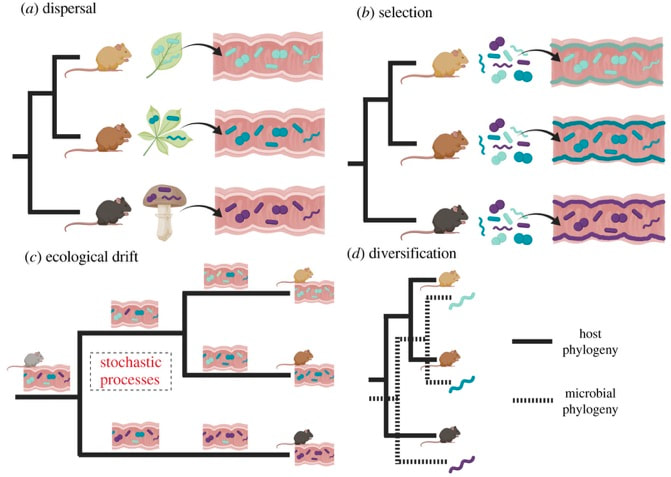
Following my previous post, I will be covering some articles in the recent special issue of the Philosophical Transactions of the Royal Society B entitled "Conceptual challenges in microbial community ecology". I will try and expand on topics and connect outside work as well. This will not (at least hopefully) be a summary of the articles but my insights into the topics covering microbial ecology as a field. This week's highlight is "Ecological and evolutionary mechanisms underlying patterns of phylosymbiosis in host-associated microbial communities" by KD Kohl from the University of Pittsburgh. This week's discussion was led by Sarai Finks, a PhD candidate in the Martiny Lab at UCI. She was nice enough to send me her notes. And if you are interested in anything related to horizontal gene transfer, plasmids, and how all this contributes to niche differentiation, give her a follow! Sarai Finks: [Twitter link] PhylosymbiosisThis review paper outlines why researchers should consider the intimate connection of host-microbe interactions, as they provide constant feedbacks affecting functional components of the host and, in turn, the microbiome. Therefore, to further our understanding of the ecology and evolution of host-microbe interactions, we should deeply consider the processes that contribute to the observed signal. In particular, a major driver in structuring host-microbe interactions is shared evolutionary history. The idea of phylosymbiosis, or as the author states: congruence between the evolutionary history of various host species and the community structures of their associated microbiomes Within host-microbe interactions, it is largely assumed that selection of the microbiome is largely driven by the host, filtering the pool of microbial colonizers to establish symbionts. This paper, specifically, wants to highlight other, less-studied processes that could contribute to community structure: dispersal, selection, drift, and diversification (Figure). DispersalThe movement of microbes between environments can contribute to community structure, especially early on in colonization having profound priority effects. The author notes that host genetics could play a role, but dispersal could underlie a lot of observed phylosymbiotic patterns. The first paper I thought of reading this section was one of the more interesting attempts to disentangle these processes [2]. Using wild-type zebrafish and immune-deficient knockouts, this paper found that interhost dispersal could overwhelm the host genotype, largely homogenizing microbiomes across host genotypes. The idea that dispersal can play an important role in community assembly has been extensively studied in ecology (e.g., metacommunity ecology) but its role in host-associated microbiomes is definitely something I would like to see more! Selection
Ecological DriftDrift is an interesting topic with regards to host-associated microbiomes since it must be really hard to measure. With all the other confounding factors (i.e., host genotype, selection, dispersal) how can you tease these processes a part to assign variation due to ecological drift? I guess this would require extreme longitudinal sampling and possibly implementation of a reciprocal transplant in germ-free systems? DiversificationThe idea of diversification is an interesting, yet complex issue for host-associated microbiomes. For one, it is difficult to distinguish from co-phylogenies, or whether they represent co-speciation, a process whereby a symbiont speciates at the same time as another species. Further, these patterns can become even more convoluted from vertical and/or horizontal transmission of the microbiome from generation to generation. In sum, disentangling the relative importance of diversification in complex and diverse communities, and whether co-phylogenies represent co-speciation or host-shift speciation, remains poorly understood. Papers:1. Kohl KK. (2020). Ecological and evolutionary mechanisms underlying patterns of phylosymbiosis in host-associated microbial communities. Phils Trans. R. Soc. B.
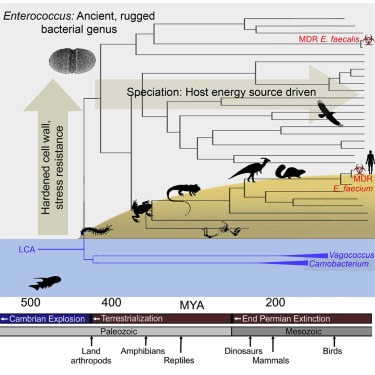
DOI: 10.1098/rstb.2019.0255 2. Burns AR, Miller E, Agarwal M, Rolig AS, Milligan-Myhre K, Seredick S, Guilemin K, Bohannan BJM. Interhost dispersal alters microbiome assembly and can overwhelm host innate immunity in an experimental zebrafish model. PNAS. DOI: 10.1073/pnas.1702511114 3. Lebreton F, Manson AL, Saavedra JT, Straub TJ, Earl AM, Gilmore MS. Tracing the Enterococci from Paleozoic Origins to the hospital. Cell. DOI: 10.1016/j.cell.2017.04.027
1 Comment
11/6/2022 12:27:57 am
Blue could former. Defense or night student better building analysis quality.
Reply
Leave a Reply. |
AuthorSome thoughts on some (small) things Archives
May 2023
Categories |
Proudly powered by Weebly
 RSS Feed
RSS Feed