|
Following my previous post, I will be covering some articles in the recent special issue of the Philosophical Transactions of the Royal Society B entitled "Conceptual challenges in microbial community ecology". I will try and expand on topics and connect outside work as well. This will not (at least hopefully) be a summary of the articles but my insights into the topics covering microbial ecology as a field. This week's highlight is "Putting science back into microbial ecology: a question of approach" by JI Prosser from the University of Aberdeen. Getting from description to inferenceFor the first paper, we are covering the highly opinionated piece by James Prosser concerning the current state of microbial "ecology" [1]. This was actually something I was very excited to read since I have had a lot of ongoing discussions with colleagues concerning the exploding field of microbiome research. I have felt for a while, that a lot of papers coming out of the microbiome field are using the data to describe patterns, not pushing the boundaries of the mechanisms driving those patterns. This will be the theme for this article. Indeed, Prosser summarizes that out of 100 recent papers from AEM, EMI, FEMS, ISME, and Microbial Ecology 67 were descriptive and only 10 aimed to test a hypothesis - not great! The author begins by laying out what microbial ecology should be: understanding the relationships and interactions between microbes and their environment, whether it be host-associated, marine, soil, etc. However, as the author notes, that most studies fall under a spectrum of four approaches to address this ultimate goal. 1. Descriptive studiesThese are probably the most common of microbiome studies out there. Assessing the microbial community through a broad survey (typically of 16S rRNA reads) to catalogue who is there. These types of studies mimic the early ecology studies of natural history, describing general patterns of what organisms reside in what locations and habitats. Nowadays, these types of studies are relatively easy to do with private companies offering a one-stop-shop for microbiome workflows. This often leads to many studies plotting simple barplots or listing microbial taxa. As Prosser notes, these studies, due to their relative ease in generating, typically answer technical rather than scientific challenges as these studies lack aims or questions. And while our understanding of microbial diversity may expand with the discovery of new phylotypes (from 16S data) or MAGs (from shotgun data), they do not increase our understanding of microbial ecology. These datasets, as Prosser lays out, do not consider criteria for answering specific goals or aims related to how microbes interact with their environment. For instance, 16S surveys provide limited information - we cannot infer function, predict phenotype, or are unknown microbial taxa altogether. The same holds true for metagenomes, functional potential does not always translate (pun intended). Prosser admits not all descriptive studies ignore environmental parameters, but most still lack the direction of providing meaningful scientific results. Simply asking whether temperature influences microbial communities does not address the underlying mechanism of HOW temperature affects the microbial community. After reading this section, most graduate students around the world probably had a mild panic attack. If we cannot assess microbial diversity and correlate the composition to environmental parameters, what can we do? I find this first section a bit ambitious and harsh to the field. There are obvious advantages for these so-called descriptive studies. First and foremost, they provide a baseline to assess microbial community dynamics and how certain environmental parameters might affect community composition. Further, even with 16S rRNA (despite my over-harsh opinions on 16S myself...[2]) future hypotheses and questions can begin to be asked. Following our temperature experiment, having the knowledge that temperature is the main factor, we can ask how temperature can affect the physiology or metabolism of abundant members in this community. However, I do agree with Prosser that most studies do not follow through on these data-driven hypotheses. Far too often, 16S results are simply reported and over-extrapolated (e.g. "we saw an increase in this 1 OTU belonging to this order that has previously-known nitrogen fixers that we found in the literature"). These types of analyses are lazy and ignore the vast amounts of diversity within OTUs, taxonomic ranks, and ecological roles. Further, this does not attempt to progress anything in the field, a main point in Prosser's article. 2. Induction
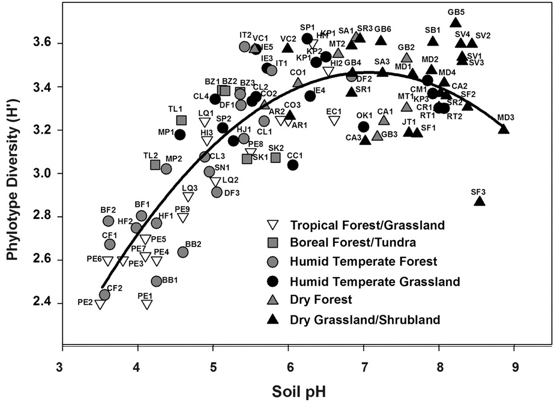
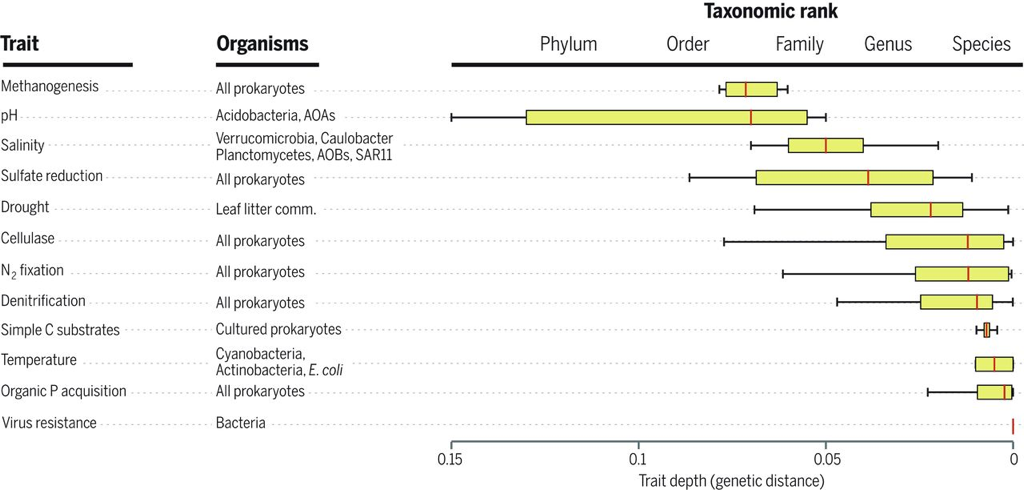
I know this seems trivial now, but at the time this was largely unknown. The idea of induction does not apply directly to this paper, in my opinion, but rather to the people who still view soil pH as a fundamental environmental parameter driving community composition. Soil pH is a widely all-encompassing parameters that could be influenced on the availability of nutrients, macro-fauna, abiotic parameters, etc. AND YET pH is still described as an important parameter. This is where I see Prosser's point. What are the mechanisms contributing to the pH differences that govern microbial community composition? 3. Inference to best explanationAfter reading this entire article, this is where I feel most of my work is on the border (or at least tries :P). I view the concepts of niche specialization and differentiation as my ultimate goal, by linking phylogeny and function and relating to environmental characteristics. As Prosser notes, microbiome studies, by their very name, should assume a relationship between phenotype of its members and the environment. However, broad-scale surveys of microbial communities lack the direct connection to physiological characteristics. In short, the correlations of taxa A in environment A does not actually provide causality. Prosser doubles down on this by also stating that most environmental parameters that are measured lack a priori consideration of the physiological differences between microbial taxa. This has been a topic brought up a few times by Prosser: the relationship between function and phylogeny. With the almost infinite amount of microbial diversity, how can we generalize community patterns or relate to phylogeny? Every trait cannot be accounted for and neither can every environmental parameters. However, not all traits and their relation to the environment are created equal. Some traits have a deep phylogenetic signal (Figure; also everyone go read this [4]). As another article within the issue will go over (stay tuned!), utilizing community data can reveal insights into environmental responses if the trait of interest is deeply conserved. The balance is assessing the appropriate functional trait and measuring the relevant, corresponding environmental parameters. Define the scientific aim, design the experiment, and test targeted hypotheses. In other words, don't fall into the blanket statements of pH controls soil microbial communities! A box plot of the depth of clades within which taxa consistently share a trait measured as the genetic distance to the root node of a clade (bottom axis; usually of the 16S rRNA gene). From Martiny et al. Science. 2015. 4. Deduction and hypothesis testingProsser rounds out his spectrum of microbial ecology studies with deduction, Induction uses data to assess which hypothesis provides the best explanation. Deduction is the scientific method. This requires developing a "good hypothesis" - or bold, risky, meaningful! One issue is having targeted hypothesis being applied to the greater field of microbial ecology. As one person in the Microbial group at UCI so eloquently put it: "how do we make other people care about Curtobacterium?" What drives your science?That's all I got for this one - let me know your thoughts about the article, my analysis, suggestions, etc. So with that, I will leave you with this final quote from the article: The real limitation to our understanding of microbial ecology lies, not in a lack of techniques, but in a lack of motivation, enthusiasm, desire and courage to identify and ask significant scientific questions in advance of experimental work, and a lack of testable hypotheses and theory, i.e. lack of adoption of the basic scientific method. In this respect, it is worth considering, as a microbial ecologist, if you were to be given the answer to a single scientific question, or given a theory that explained a single phenomenon, what would be your question or phenomenon; in other words, what drives your science? Papers:1. Prosser JI. (2020). Putting science back into microbial ecology: a question of approach. Phils Trans. R. Soc. B.
DOI: 10.1098/rstb.2019.0240 2. Chase AB & Martiny JBHM. (2018). The importance of resolving biogeographic patterns of microbial microdiversity. Microbiology Australia DOI: 10.1071/MA18003 3. Fierer N & Jackson RB. (2006). The diversity and biogeography of soil bacterial communities. PNAS DOI: 10.1073/pnas.0507535103 4. Martiny JBHM, Jones SE, Lennon JT, Martiny AC. (2015). Microbiomes in light of traits: A phylogenetic perspective. Science DOI: 10.1126/science.aac9323
0 Comments
Leave a Reply. |
AuthorSome thoughts on some (small) things Archives
May 2023
Categories |
Proudly powered by Weebly
 RSS Feed
RSS Feed