BackgroundA major goal in evolutionary biology is to understand the origin and maintenance of genetic variation and diversity. This framework allows us to identify the processes governing species distributions and diversity. In microbes, it is difficult to delineate species boundaries and identify populations undergoing diversification. Most studies employ comparative genomics by collating closely-related genomes from various databases. However, these analyses do not consider whether strains were from the same habitat and, thus, we cannot examine whether they are a part of the same populations of interacting genotypes. Being able to compare diverging populations allows us to identify the mechanisms contributing to microbial speciation and diversification. In theory, speciation is dependent on 1) selection leading to ecological differentiation and 2) barriers to recombination. This is similar to sexual organisms; however, the lack of sexual reproduction in microbes makes it harder to differentiate. These are all topics that fascinate me in microbial ecology and evolution. To this day (for a variety of reasons I will try and cover in another post), we still grapple with the mechanisms driving microbial diversification and speciation. The case of Vibrio provides an excellent story addressing these topics and was one of the more interesting studies that motivated me to pursue population dynamics in environmental microbes. Vibrio - a test case
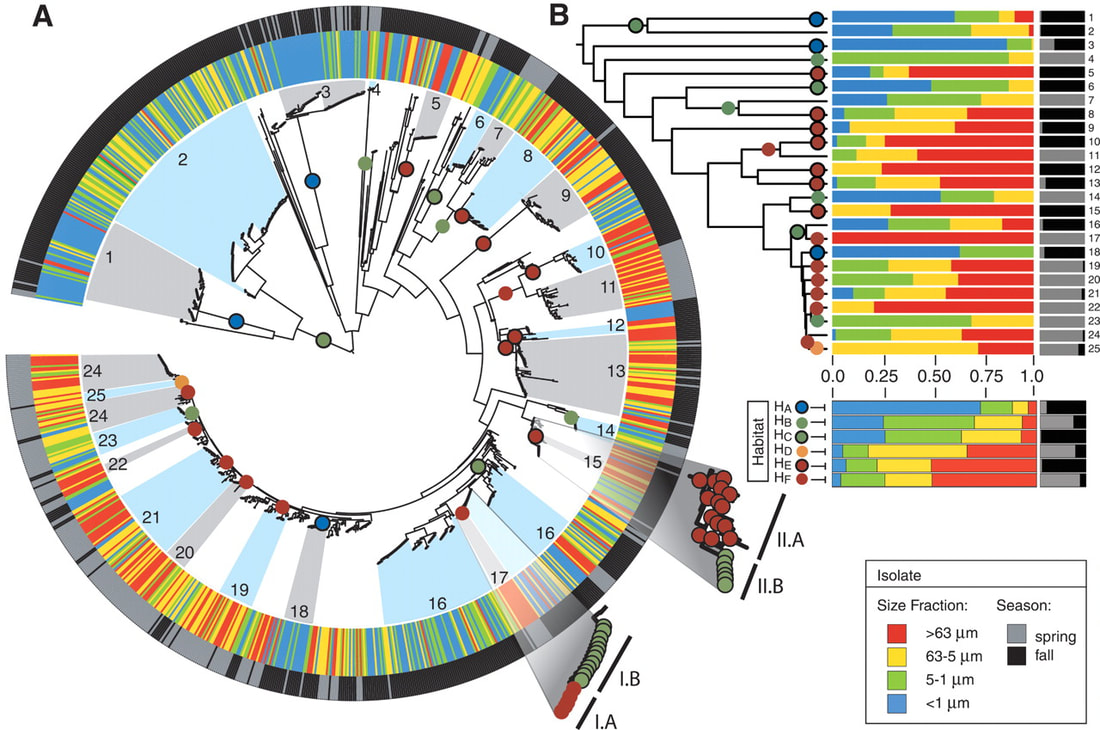
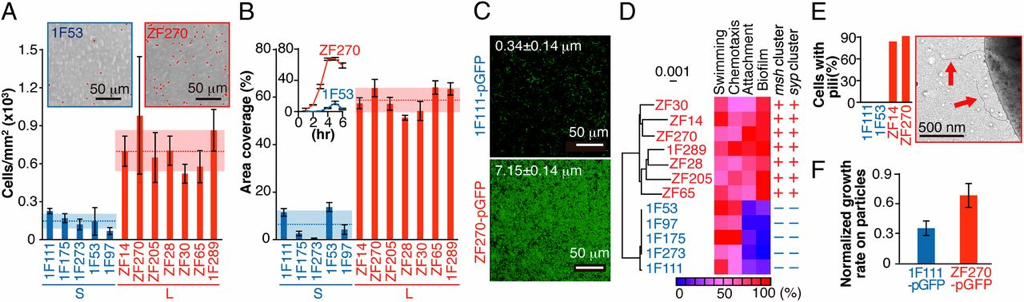
Organic particles in the ocean are thought to be hotspots for community aggregation. Insomuch, that marine particle-attached communities can undergo ecological succession, where species turnover is dictated by resource availability. In this case, motile bacteria arrive first and begin the degradation of particles and later followed by secondary consumers of the carbon byproducts [2]. Population GenomicsFollowing the observations that Vibrio strains appeared to have preferences for particle size, a study lead by Shapiro sought to identify the genomic mechanisms of divergence potentially leading to bacterial speciation [3]. Utilizing 20 genomes from large (L) or small (S) particles sizes (proxy for habitat), they first found evidence for differentiation among closely-related strains. For perspective, the strains all shared identical 16S rRNA genes and >99% average amino acid identity across the genome. The relevant differences among these strains resided in 725 "ecoSNPs" that cluster in genomic space in 11 concentrated regions (see below figure). These ecoSNPs supported habitat divergence, while the rest of the genomic SNPs provide incongruence with the ecological split into distinct habitats. Further, these SNPs contain low within-habitat diversity suggesting these regions are a result from recent recombination and have subsequently swept through the populations, coined gene-specific sweeps. Analyses of the core and flexible genome support these conclusions as they identified core regions more frequently recombined within-habitat than between-habitat. A) core genome phylogeny for chromosome I and II B) genome regions showing localization of ecoSNPs Ecological DifferentiationEvidence for high recombination within-habitat strains led the authors to inquiry how these populations may be differentiating. Due to the apparent absence of geographic barriers, the sympatric populations must be differentiating due to ecological differentiation [4], in this case, habitat preference. Indeed, the flexible genome provided evidence as L-population strains encoded flexible genes related to biofilm formation and the biosynthesis of MSHA for chitin adhesion. Thus, these closely-related strains are partitioning their microenvironments between particle-associated or free-living (migratory) populations. The acquisition of habitat-specific flexible genes can lead to ecological specialization, which further depresses recombination between the incipient populations. The question then becomes, can microscale environmental heterogeneity in microbial communities allow for the coexistence of closely-related organisms? Or put another way, are behavioral adaptations to particle-associated vs. free-living lifestyles strong enough to create boundaries to gene flow and structure population divergence? A follow-up study examined strains from both populations for various phenotypic measurements related to particle-association, including swimming speeds, cell sizes, flagellation, and chemotaxis [5]. Initially, strains from the L- population exhibited a differential ability to attach to particles (agarose, cellulose, alginate) and form biofilms (see below figure). However, S-populations strains (migratory, free-living) did not outcompete L-populations strains in swim speed nor chemotaxis in a steady resource gradient microfluidic chamber. Instead, the S-population strains, under time-varying conditions, were able to migrate to new nutrient supplies (really cool microfluidic videos here). Together, these closely-related strains appear to differentiate on fine-scale behavioral adaptations. Phenotypic assays for S- and L-populations. A) Attachment to polystyrene. B) Biofilm formation and C) images. D) Correlation between S and L for phenotypic traits and genes. E) Number of pili F) Growth rate on alginate particles Papers1. Hunt DE, David LA, Gevers D, Preheim SP, Alm EJ, Polz MF. (2008). Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science 320: 1081–1085.
2. Datta MS, Sliwerska E, Gore J, Polz MF, Cordero OX. (2016). Microbial interactions lead to rapid micro-scale successions on model marine particles. Nature Communications 7: 11965. 3. Shapiro BJ, Friedman J, Cordero OX, Preheim SP, Timberlake SC, Szabó G, Polz MF, Alm EJ (2012). Population genomics of early events in the ecological differentiation of bacteria. Science 336: 48–51. 4. Cordero OX, Polz MF. (2014). Explaining microbial genomic diversity in light of evolutionary ecology. Nature Reviews Microbiology 12: 263–273. 5. Yawata Y, Cordero OX, Menolascina F, Hehemann JH, Polz MF, Stocker R. (2014). Competition–dispersal tradeoff ecologically differentiates recently speciated marine bacterioplankton populations. Proceedings to the National Academy of Sciences 111: 5622–5627.
0 Comments
Leave a Reply. |
AuthorSome thoughts on some (small) things Archives
May 2023
Categories |
Proudly powered by Weebly
 RSS Feed
RSS Feed