Theme of the month: horizontal gene transferHGT has the power to accelerate evolution by introducing novel alleles across phylogenetically distant taxa. At the same time, rampant HGT can blur species boundaries and is expected to result in a mosaic of genes. I have always been fascinated by the idea that bacteria undergo rampant HGT insomuch that there have been previous proposals for a "web of life". As much as HGT can contribute to rapid diversification, there is also strong evidence for cohesive bacterial lineages, which is fundamental to our understanding of biodiversity in the microbial kingdom. These ideas seem to create an evolutionary "tug-of-war" between local adaptation mediated via acquisition of beneficial alleles and shared phylogenetic history. Here, I just wanted to highlight some new(ish), excellent papers addressing the idea of HGT across multiple scales, from theoretical to empirical, and HGT across kingdoms. Brief Introduction: HGT vs. recombination
At the same time, there are interesting patterns when looking at closely-related bacteria. For one, when comparing genomes of closely-related bacteria, many of the genes in the genomes are unique, or not found in all strains. These genes are collectively referred to the pan-genome. Repeatedly, studies find almost infinite pan-genomes when surveying bacterial groups. Whether these genes are neutral or are a result of frequent adaptive HGT in local environments remains up for debate. Some have proposed that local populations can tap into a shared gene pool as many of these accessory or flexible genes are under frequency-dependent selection [2]. This would suggest that flexible genes are under strong environmental selective pressures. While HGT can possibly provide beneficial fitness effects, there must be a cost to acquiring foreign DNA. Simply put, for adaptive genes to be maintained in a population, the beneficial effects must far out-weigh detrimental effects or genetic drift [3]. This is why HGT fascinates me. There is an intrinsic cost to accepting foreign DNA, but we know HGT can drastically shape microbial evolution. Personally, I believe many studies have overestimated HGT events or are documenting "evolutionary relic" events, such as using gene homology to overestimate the extent of HGT in bacterial genomes. This is why, I suggest when reading the following list to keep in mind some questions: 1. How prevalent is HGT in natural systems? Obviously we can induce genetic exchange in the laboratory or under high selective pressures, but how common are these events in nature? 2. Is HGT a hand-wavy explanation for unexplained genomic patterns? By comparing disparate genomes from across different environments we may be overestimating HGT events and masking the local adaptation of microbial populations. That's enough of my rambling, let's get to the good stuff: Some interesting modes of HGT transferLet's start with phages.
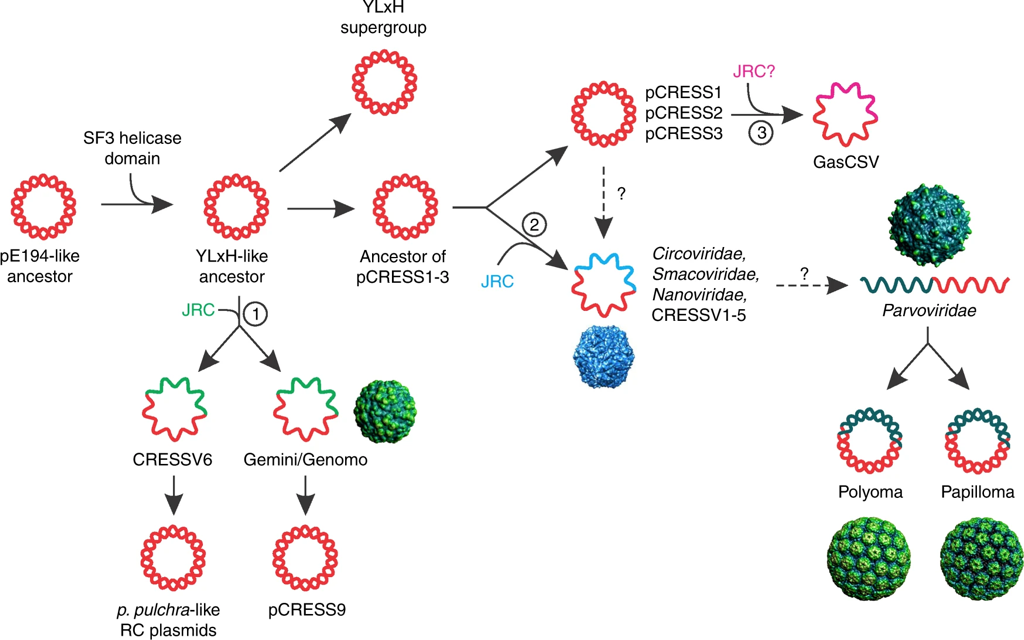
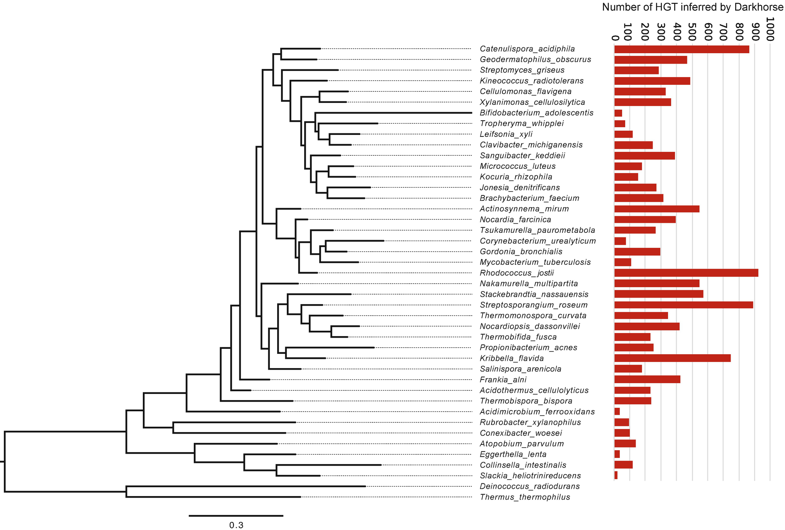
Probably the most studied form of HGT is antibiotic resistance. A couple years ago, a paper from a French research group addressed the acquisition of antibiotic resistance genes mediated through phages. Specifically, this paper addressed the issue in antibiotic resistance gene detection. Inevitably, they find most detected resistance genes were overestimated ("inflated false positives") and that phages rarely encode genes related to antibiotic resistance [5]. The contrast of these first two papers is great! Phages can obviously mediate HGT but the genes being distributed are highly variable. A small shift in gears now. I want to keep discussing phages but move on to the origin of phages. So, for the next paper, we will look at the origin of single-stranded DNA viruses from traditionally exchanged plasmids in bacteria and archaea. ssDNA viruses replicate via the Rep protein of the HUH endonuclease family, a mechanism also found in plasmids. By exploring the relationships among Rep-encoding DNA viruses and transposons from plasmids, the researchers conclude that the origins of ssDNA viruses can be traced to prokaryotic plasmids [6]. Next up, plamids! Speaking of plasmids, a recent paper documents the presence of a large megaplasmid, with the shared genetic potential to replicate, transcribe, and repair DNA as another closely-related megaplasmid [7]. Megaplasmids, themselves, are evolutionary interesting as the maintenance of these large extra-chromosomal regions possess greater evolutionary costs. The two analyzed in this study suggest strong selective pressures to maintain genetic synteny; while high divergence between orthologous groups suggest independent evolution from a common ancestral plasmid. HGT vectors such as plasmids are undergoing evolutionary processes themselves - they're not just vectors! Cross-kingdom HGTBacterial operons are unique and ubiquitous. They encode the transcription, translation, and production of proteins all in tandem. Bacteria couple these processes by clustering genes into operons, while eukaryotes spatially and temporally separate these processes. These are fundamental differences in metabolism between kingdoms. The next paper, however, found a bacterial operon being transferred, acquired, and maintained in a fungal lineage [8]. After acquisition, the operon underwent structural changes to integrate into eukaryotic synthesis - crazy! The encoded siderophore cluster maintained its high gene clustering in the fungal genome while being modified through transcription including polyadenylation. This paper highlights the boundaries of cross-domain gene transfer for the integration of a complex metabolic pathway. Detection of HGT eventsLastly, I just want to finish with some interesting reports into the frequency of HGT in bacterial systems, specifically in the Actinobacteria phyla. A recent edition on HGT included a detailed account of how HGT can shape evolution in Actinobacteria [9]. These examples include overviews of Streptomyces and Salinispora (very new and dear to me now). In both cases, the authors detail the exchange of large contiguous biosynthetic gene clusters (BGCs) and their relation to HGT, specifically a "plug-and-play" model of evolution where BGCs can be swapped in and out in concentrated genomic islands. To me, this seems extraordinary as HGT should come with (mostly) deleterious fitness costs and the integration of large genomic segments (>30kbp!!!) is hard to wrap my head around. Further, an analysis from the authors inferring HGT events in the Actinobacteria phylum is astounding (below figure), but part of me questions whether this is far overestimating HGT events. Or are we far too liberal with what we classify HGT events? For instance, a paper a couple years ago found that HGT events in Streptomyces were actually quite rare, on the order of 10 genes per million years were acquired and maintained [10]. This would make the transfer of entire BGCs almost unheard of! I would love to hear everyone else's perspective on this front. Again, I am blown away with HGT and really interested in the topic. There are tons and tons of papers I cannot even begin to break down, so let me know your thoughts! Papers:1. Rocha EPC. (2018). Neutral Theory, microbial practice: challenges in bacterial population genetics. Molecular Biology and Evolution 35: 1338-1347.
2. Polz MF, Alm EJ, Hanage WP. (2013). Horizontal gene transfer and the evolution of bacterial and archaeal population structure. Trends in Genetics 29: 170-175. 3. Baltrus DA. (2013). Exploring the costs of horizontal gene transfer. Trends in Ecology and Evolution. 28: 489-495. 4. Frazão N, Sousa A, Lässig M, Gordo I. (2019). Horizontal gene transfer overrides mutation in Escherichia coli colonizing the mammalian gut. Proceedings to the National Academy of Sciences 116(36): 17906-17915. 5. Enault F, Briet A, Bouteille L, Roux S, Sullivan MB, Petit MA. (2017). Phages rarely encode antibiotic resistance genes: a cautionary tale for virome analyses. The ISME Journal 11: 237-247. 6. Kazlauskas D, Varsani A, Koonin EV, Krupovic M. (2019). Multiple origins of prokaryotic and eukaryotic single-stranded DNA viruses from bacterial and archaeal plasmids. Nature Communications 10:3425. 7. Smith BA, Leligdon C, Baltrus DA. (2019). Just the two of us? A family of Pseudomonas megaplasmids offers a rare glimpse into the evolution of large mobile elements. Genome Biology Evolution 11(4): 1223-1234. 8. Kominek J, Doering DT, Opulente DA, Shen XX, Zhou X, DeVirgilio J, Hulfachor AB, Groenewald M, Mcgee MA, Karlen SD, Kurtzman CP, Rokas A, Hittinger CT. (2019). Eukaryotic acquisition of a bacterial operon. Cell 176: 1356-1366. 9. Park CJ, Smith JT, Andam CP. (2019). Horizontal gene transfer and genome evolution in the phylum Actinobacteria. In: Villa T., Viñas M. (eds) Horizontal Gene Transfer. Springer, Cham 10. McDonald BR, Currie CR. (2017). Lateral gene transfer dynamics in the ancient bacterial genus Streptomyces. mBio 8: e00644-17.
0 Comments
Leave a Reply. |
AuthorSome thoughts on some (small) things Archives
May 2023
Categories |
Proudly powered by Weebly
 RSS Feed
RSS Feed