Introduction to microbial biogeographyUtilizing a biogeographic framework is extremely powerful as biologists can understand the ecological and environmental factors driving the distribution of taxa. In addition, extensions of this include phylogeography where evolutionary biologists can understand barriers to dispersal and geological events that may contribute to species' distributions. More recently, these patterns have been observed for microbes. However, microbial biogeographic patterns are typically associated with environmental factors at the community level. This is in stark contrast to the traditional work in plants and animals, where species' distributions and/or populations are described. Previously, I wrote a commentary with my PhD advisor, Jennifer Martiny, going over some of these ideas [1]. Recently, I have seen a lot more papers moving beyond community-level patterns and trying to identify the ecological and evolutionary processes structuring the distribution of microbial taxa. So, for this month, I thought everyone could take a deep dive into some of these amazing papers. I tried to find papers referencing soil, marine, and host-associated bacteria to give a little bit of something for everyone. This list is in no way inclusive, so please feel free to share other papers you have seen as well! I will be more than happy to add to this list. Global-level patternsFirst couple of papers I want to highlight are looking at the global distribution of microbial taxa. To start, we can look at soil bacteria. A lot of papers have previously shown that, at global levels, various edaphic properties can structure soil communities, including pH, temperature, and salinity. Greenlon et al. examined the phylogenetic diversity of the nodule-forming genus, Mesorhizobium, by sampling chickpeas across various soil types, climates, continents, etc. [2].
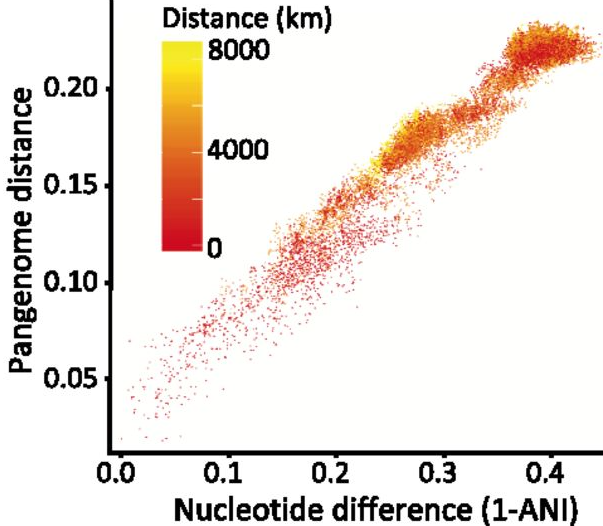
The second paper dives into maybe the most abundant organism on the planet, the SAR11 clade. This order has a vast global distribution in oceans with finer clades exhibiting differential geographic patterns. Despite its ubiquity and global distribution, SAR11 remains incredibly difficult to culture and also creates problems reconstructing MAGs from environmental surveys. As such, this next paper attempts to investigate the genomic diversity of the SAR11 clade using read recruitment of oceanic samples to identify single-amino acid variants [3]. One issue I keep coming across in any investigation into the SAR11 clade is that most studies are examining genetic diversity across an entire order. Is it really surprising that different families, genera, species, populations (if we can get to this level) within the SAR11 order have differential distributions? And why is it applicable to then look at non-synonymous variation? In any case, the authors further extrapolate amino acid variants to physico-chemical properties to conclude that the hydrophobic interactions of amino acids are under purifying selection. Another question I had how many amino acid positions would be affected by the chemistry, is this restricted to the active sites? How did they tease a part neutral divergence in these proteins across divergent taxa? Finally, the paper concludes that proteotypes within a SAR11 subclade exhibit differential biogeographic patterns with temperature being a driving determinant of spatial partitioning. But then they conclude that "lending credence to the idea for marine systems that 'everything is everywhere but the environment selects'"? - shouldn't these distinct biogeographic patterns reflect the opposite? Regional biogeographic patternsRates of homologous recombination between closely-related strains have largely been inferred from genome sequences, but Potnis et al. utilize in vitro recombinant strains along with wild-type isolates to determine the extent of HR [4]. The authors identified large recombining fragments (upwards of 30kbp) in concentrated genomic islands. Functional annotations of these regions revealed genes related to the transition of pathogenic lifestyles of the plant pathogen.
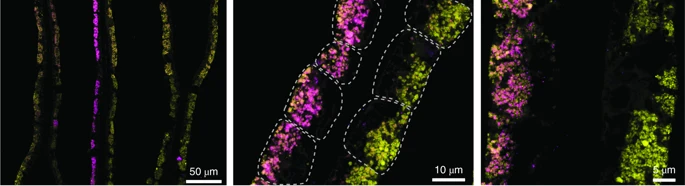
Fine spatial scaleStaying with soil dynamics, next I want to look at this really neat paper that sampled Streptomyces strains at spatial microscales (mm to cm). Similarly to the SAR11 clade, this "genus" encompasses extensive genetic diversity. Based on these ambiguous delineations, I think a lot of studies have overestimated the extent of HGT in structuring Streptomyces diversity; actually a recent study quantified this showing they are indeed rare HGT events across distantly-related Strepto lineages. Back to this paper which isolated 32 strains from soil aggregates that shared >98.6% ANI [6]. Despite this, conspecific strains harbored extensive gene content differences in the flexible genome and proposed that the massive gene fluxes are probably mediated by Actino ICEs (AICEs). Some of these indels included biosynthetic gene clusters (BGCs) - this might only be a cool result for me! In conclusion, the observed gene fluxes between microscale spatially-separated strains was mediated by frequent transfer among closely-related strains via conjugation. This last paper is really interesting as **SPOILER** they show as many as 16?!?!? strains of intracellular symbionts can coexist in a single individual of deep sea mussels [7]. I find this result fascinating since we would expect highly similar strains to outcompete one another via the competitive exclusion principle. In this system, it was thought the mussels hosted two intracellular symbionts, one for sulfur-oxidizing (SOX) and another for methane-oxidizing - both contribute energy sources for carbon fixation. By 16S rRNA standards, each mussel host was thought to harbor a single symbiont species of SOX and methane oxidizers. This paper utilized metagenomes and metatranscriptomes to identify variable strain diversity in a single mussel host. Further, these variations were linked to strain-specific genes related to key functional processes in the SOX symbionts. For instance, some symbiont strains expressed genes related to H2 oxidation; the authors also confirmed with imaging analysis (below). This imaging revealed such fine-scale spatial partitioning that it was restricted to the host bacteriocytes - that is incredible compartmentalization! Simultaneous FISH of hydrogenase operon (violet) and 16S rRNA (green) of the SOX symbiont in gill tissue of B. azoricus from site encoding genes related to H2 oxidation. Papers:1. Chase AB, Martiny JBH. (2018). The importance of resolving biogeographic patterns of microbial microdiversity. Microbiology Australia 39(1): 5-8.
2. Greenlon A, Chang PL, Damtew ZM, Muleta A, Carrasquilla-Garcia N, Kim D, Nguyen HP, Suryawanshi V, Krieg CP, Yadav SM, Patel JS, Mukherjee A, Udupa S, Benjelloun, Thami-Alami I, Yasin M, Patil B, Singh S, Sarma BK, von Wettberg EJB, Kahraman A, Bukun B, Assefa F, Tesfaye K, Fikre A, Cook DR. (2019). Global-level population genomics reveals differential effects of geography and phylogeny on horizontal gene transfer in soil bacteria. Proceedings to the National Academy of Sciences 116(30): 15200-15209. 3. Delmont TO, Kiefl E, Kilinc O, Esen OC, Uysal I, Rappe MS, Giovannoni S, Eren AM. (2019). Single-amino acid variants reveal evolutionary processes that shape the biogeography of a global SAR11 subclade. eLife 8:e46497. 4. Potnis N, Kandel PP, Merfa MV, Retchless AC, Parker JK, Stenger DC, Almeida RPP, Bergsma-Vlami M, Westenberg M, Cobine PA, de la Fuente L. (2019). Patterns of inter- and intrasubspecific homologous recombination inform eco-evolutionary dynamics of Xylella fastidosa. The ISME Journal 13: 2319–2333. 5. Chase AB, Arevalo P, Brodie EL, Polz MF, Karaoz U, Martiny JBH. (2019). Maintenance of sympatric and allopatric populations in free-living terrestrial bacteria. mBio 10(5): e02361-19. 6. Tidjani A, Lorenzi J, Toussaint M, van Dijk E, Naquin D, Lespinet O, Bontemps C, Leblond P. (2019). Massive gene flux drives genomic diversity between sympatric Streptomyces conspecifics. mBio 10(5): e01533-19. 7. Ansorge R, Romano S, Sayavedra L, Porras MAG, Kupczok A, Tegetmeyer HE, Dubilier N, Peterson J. (2019). Functional diversity enables multiple symbiont strains to coexist in deep-sea mussels. Nature Microbiology
0 Comments
Leave a Reply. |
AuthorSome thoughts on some (small) things Archives
May 2023
Categories |
Proudly powered by Weebly

 RSS Feed
RSS Feed