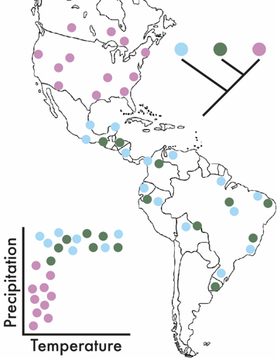
This is where adapting a biogeographic framework can be powerful. It allows us to assess the processes controlling the geographic distribution of species over space and time. It can incorporate ecological and environmental factors (e.g., temperature and precipitation) while also allowing for the integration of phylogeography to assess evolutionary processes (Figure). These ideas have a rich history in plant and animal communities and have lead to the develop of ground-breaking ecological theories attempting to explain species diversity and distribution (for some ecological background, see MacArthur-Wilson, Hubbell's Neutral Theory, and/or a nice synthesis by M Vellend). In the past two decades, these patterns have largely been explored in microbial communities, reflecting the importance of selection of environmental conditions based on correlations between microbial composition and the environment. For instance, one of the first papers to really explore these patterns showed soil bacterial communities were highly influenced by pH [1]. However, there is a large disconnect between theoretical and empirical work conducted in plants and animals to microbes, as the former are studied at the species level and describe large-scale patterns of species’ distributions. By concentrating on finer-genetic resolutions (i.e., species and population levels) we can better detect the eco/evo processes contributing to the maintenance of microbial diversity [2]. Deterministic or Stochastic?!?!For this section, I just want to highlight some processes that have been shown (in either bacteria or macroorganisms) to maintain species diversity. For the ease of this blog post, I will broadly separate these into two major categories, deterministic and stochastic processes. This, in no way comprehensive list is mainly derived from the amazing ecology grad course taught by my PhD advisor, Jennifer Martiny at UC Irvine.
This is where I think microbial ecologists can vastly expand our understanding of the tremendous microbial diversity. For instance, niche partitioning requires relating traits to phylogeny which should reflect differential environmental distributions. Often, this requires quantifying traits of closely-related bacteria as the competitive exclusion principle predicts a limitation to niche differences. Furthermore, huge efforts are being made to quantify the relative impacts of deterministic v. stochastic processes in microbial studies, both from theoretical frameworks [3] to experimental [4]. Papers:1. Fierer N & Jackson RB. (2006). The diversity and biogeography of soil bacterial communities. Proceedings to the National Academy of Sciences 103(3): 626-631.
2. Chase AB & Martiny JBH. (2018). The importance of resolving biogeographic patterns of microbial microdiversity. Microbiology Australia 39(1): 5-8. 3. Ning D, Deng Y, Tiedje JM, Zhou J. (2019). A general framework for quantitatively assessing ecological stochasticity. Proceedings to the National Academy of Sciences 116(34): 16892-16898. 4. Albright MBN, Chase AB, Martiny JBH. (2019). Experimental evidence that stochasticity contributes to bacterial composition and functioning in a decomposer community. mBio 10:e00568-19.
0 Comments
Leave a Reply. |
AuthorSome thoughts on some (small) things Archives
May 2023
Categories |
Proudly powered by Weebly
 RSS Feed
RSS Feed