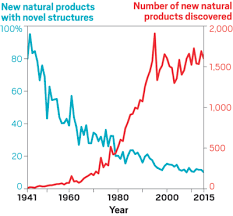
Natural ProductsAbout 9 months ago, I started my postdoc in a brand new field, natural products. Instead of focusing on the processes contributing to microbial diversity and diversification, the objective is to utilize microbes for the discovery of new natural products. As a total novice, I was blown away by the amazing work being done to quantify single microbial compounds. So, as I look back on what I learned over the past few months, I just wanted to highlight some of the amazing and novel techniques being used to give us insights into the structural diversity and complexity of natural products. This is my (very) humble understanding of natural products and definitely going to be a bit biased towards Scripps and marine sediment bacterium!
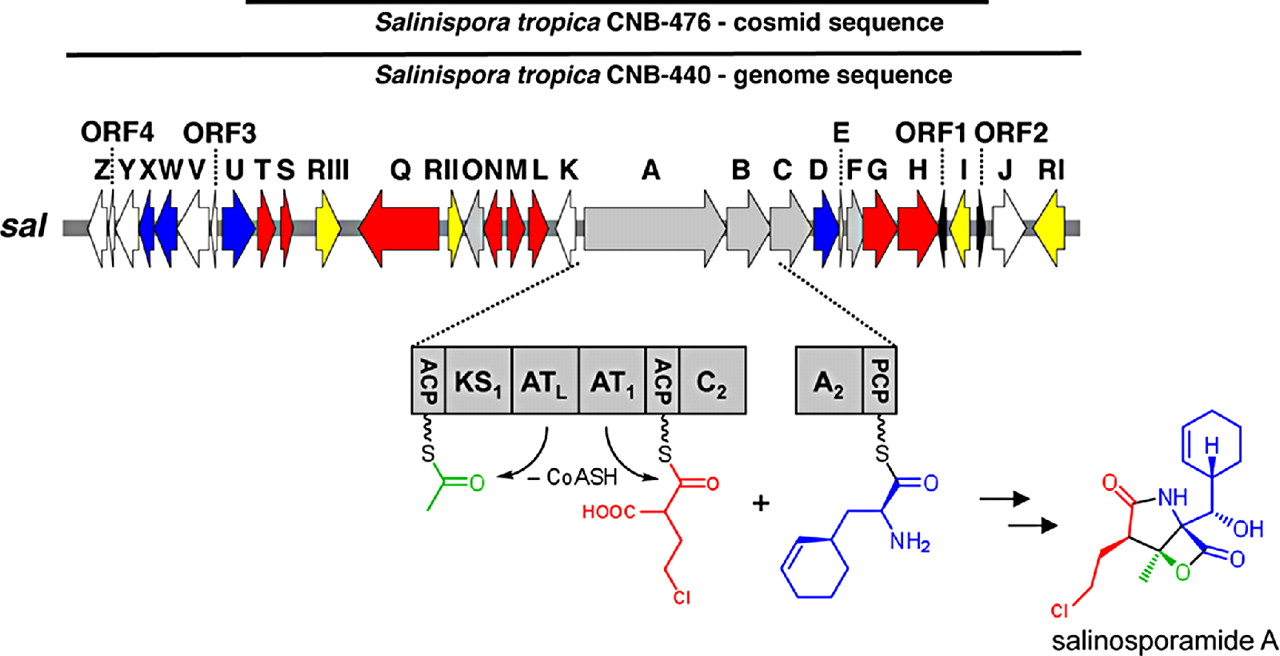
Genome Mining for NPsOf course I would start here! Obviously, the major advances in sequencing technology and costs have allowed for extensive mining of genomes for genetic signatures related to the production of natural products. In bacteria, these genes are typically clustered in the genome to build proteins in a modular fashion (akin to a car assembly line). Modules within these biosynthetic gene clusters (BGCs) enable the loading, attachment, and extension of building blocks to produce NPs (below Figure).
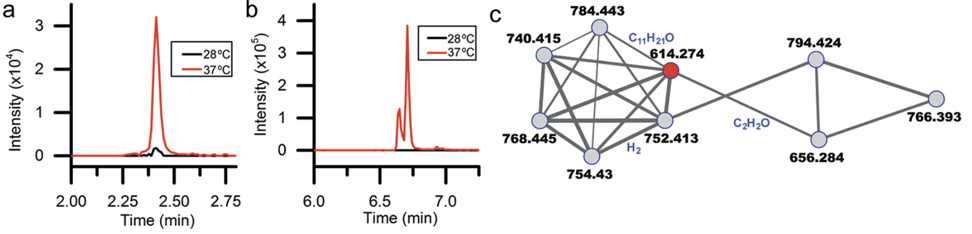
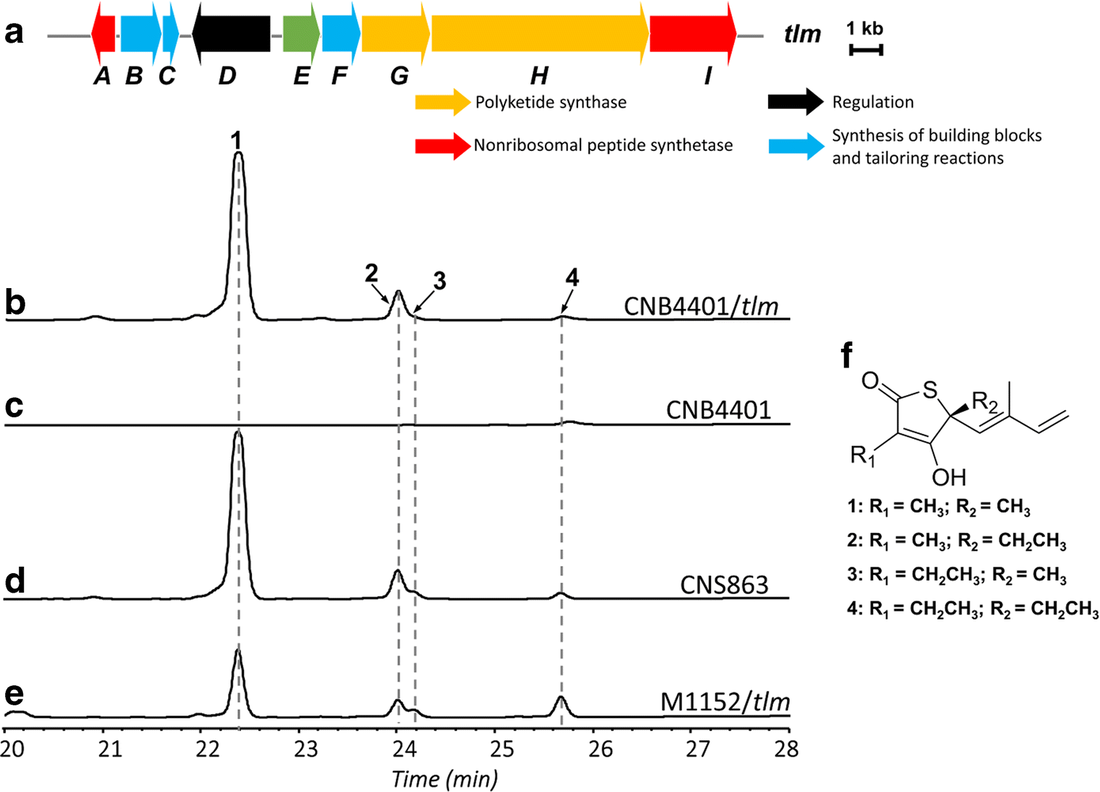
Decades of work identifying novel genomic signatures for BGCs has allowed for extensive surveys across bacterial taxa. Most notably are the Actinobacteria, which include the well-studied NP producing genera such as Streptomyces and Salinispora. Genome mining has revealed large portions of the genome can be dedicated to the production of NPs. This computational approach assesses the entire biosynthetic potential of an organism rather than examining individual metabolites (of which may or may not be expressed in culture conditions due to regulation or environmental signaling). Mass SpectrometryTraditionally, strains are grown in culture and crude extracts are examined to determine which products a bacteria might be producing. This led to the one strain many compounds (OSMAC) approach to characterize diverse compounds in different culture conditions (see below figure). However, analyzing these crude extracts are difficult as secondary metabolites are highly diverse in their size, structure, and physicochemical properties. Instruments such as the LC-MS (liquid chromatography - mass spectrometry) can separate and identify masses of compounds, but still an organisms can produce hundreds of compounds in any given sample. Much work is still to be done, but analytical tools, such as GNPS and MZMine, can aid with the data processing and identification (and dereplication) to characterize compounds. Identification of molecules A) 614.27 m/z and B) 754.44 m/z in high temperature culturing conditions. C) Molecular networking based on MS2 spectra (via GNPS) clustered both masses with a known natural product (red node) [3]. Heterologous Expression of BGCsLinking the identification of BGCs to their products using mass spec is really difficult. As such, most of the "low-hanging fruit" have been characterized in most model organisms for natural products research. This has led to some creative approaches to identify novel secondary metabolites. With the advances in computational tools, exploring biosynthetic potential in the genome has revealed a number of "orphan" BGCs in genomes, or identified BGCs that have yet to be linked to its corresponding molecule.
Papers:1. Pye CR, Bertin MJ, Lokey RS, Gerwick WH, Linington RG. (2017). Retrospective analysis of natural products provides insights for future discovery trends. Proceedings to the National Academy of Sciences 114(22): 5601-5606.
2. Eustáquio AS, McGlinchey RP, Liu Y, Hazzard C, Beer LL, Florova G, Alhamadsheh MM, Lechner A, Kale AJ, Kobayashi Y, Reynolds KA, Moore BS. (2009). Biosynthesis of the salinosporamide A polyketide synthase substrate chloroethylmalonyl-coenzyme A from S-adenosyl-l-methionine. Proceedings to the National Academy of Sciences 106(30): 12295-12300. 3. Sidebottom AM, Johnson AR, Karty JA, Trader DJ, Carlson EE. (2013). Integrated metabolomics approach facilitates discovery of an unpredicted natural product suite from Streptomyces coelicolor M145. ACS Chemical Biology 8: 2009-2016. 4. Zhang JJ, Moore BS, Tang X. (2018). Engineering Salinispora tropica for heterologous expression of natural product biosynthetic gene clusters. Applied Microbiology and Biotechnology 102(19): 8437-8446.
1 Comment
|
AuthorSome thoughts on some (small) things Archives
May 2023
Categories |
Proudly powered by Weebly
 RSS Feed
RSS Feed