|
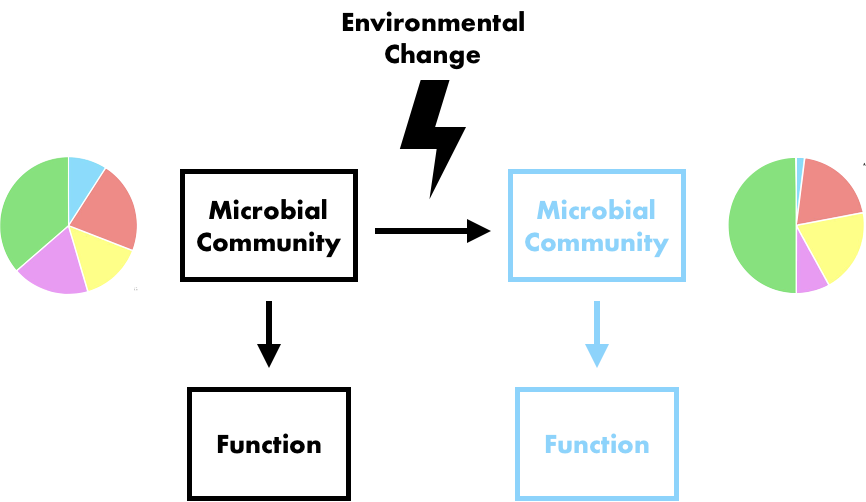
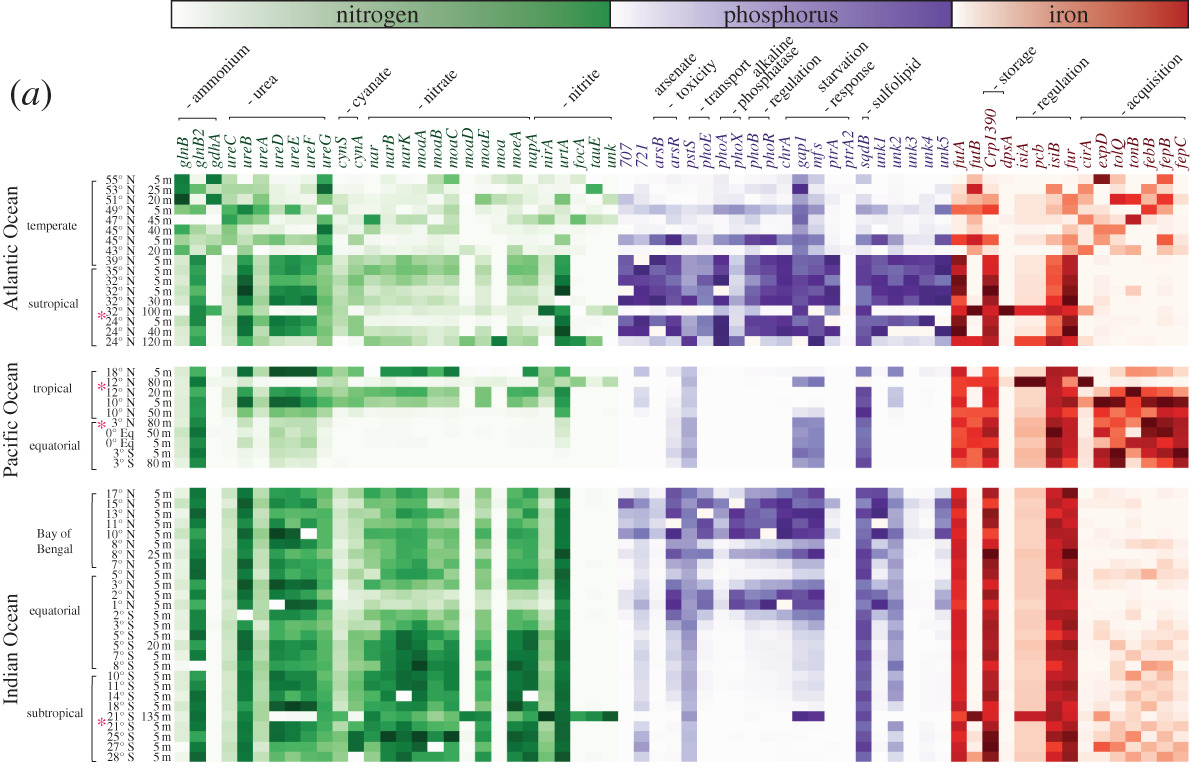
Following my previous post, I will be covering some articles in the recent special issue of the Philosophical Transactions of the Royal Society B entitled "Conceptual challenges in microbial community ecology". I will try and expand on topics and connect outside work as well. This will not (at least hopefully) be a summary of the articles but my insights into the topics covering microbial ecology as a field. And if you want to read along, that previous post contains the schedule of the papers for each week. This week's highlight is "Linking regional shifts in microbial genome adaptation with surface ocean biogeochemistry" by CA Garcia, GI Hagstrom, AA Larkin, LJ Ustick, SA Levin, MW Lomas, and AC Martiny from the University of California, Irvine. Using 'omics to inform biogeochemical cyclesIn any environmental (or even host-associated) microbiome study, the typical ultimate goal is to relate the patterns, observations, and mechanisms driving microbial composition to ecosystem processes. This allows for predictions on how variation in abiotic factors (e.g., global climate change) will affect global biogeochemical cycles. This is not a trivial task as it requires understanding how the microbial community (and its relevant members) might change to the abiotic factors and how the resulting function will be affected (Figure). This framework is extremely powerful as it allows a predictive approach by considering the microbial traits themselves. A focus on microbial traits enables the ability to link community dynamics, environmental responses, and ecosystem processes. However, this "trait-based framework" (as a side note, highly recommended reading [1]) is often difficult to assess because it requires knowing which taxa and/or traits are useful biomarkers contributing to ecosystem processes. In this week's paper by Garcia et al. [2], the authors examine the ratios of carbon:nitrogen:phosphorus (C:N:P) and how microbes contribute to these global carbon and nutrient cycles. Typically, nutrient quantifications in the environment are hard to discriminate; for example, how to tell the difference between N availability if it's in the form of ammonium or nitrate? Well, the authors in this paper took a different perspective and thought about using microbial traits (or the genes encoding these traits) to infer relative nutrient conditions. This is made possible by the assumption that these genes within certain key taxa (i.e., Synechococcus and Prochlorococcus) will reflect "rapid" adaptation to nutrient conditions. For instance, in N-stressed environments cells will encode for more varieties of N uptake (e.g., urea, nitrite, and nitrate assimilation). This works in this system as decades of work underlies the distribution of nutrient niche partitioning in these two sister cyanobacterial genera (for more information, you can read my previous post here about the evolution of N genes in Prochlorococcus [3]). Further, these photosynthetic microbes largely contribute to global carbon cycles. A great question becomes, in other systems, which taxa and/or which genes would be adequate biomarkers to assess to link to ecosystem processes? Biogeography of nutrient-related genesThis paper utilized a global sampling scheme across multiple oceans, latitudes, and used various technical characterizations, such as metagenomics, flow cytometry, particulate organic matter, nutrient measurements, etc. The crux of this paper hangs on reliable classification of nutrient-related genes to nitrogen, phosphorus, and iron and their taxonomic origin. Again, this really only becomes convincing due to the decades of work into this system. Using a curated reference database of the Pro and Syn nutrient-genes, the authors could mine out reads from the metagenomic data to assess the diversity and distribution of these genes across various oceanic stations. After normalizing for single-copy marker genes, the authors found that certain oceans were more or less enriched in specific nutrient genes. These gene abundances corresponded to the environmental conditions, suggesting that the presence/absence of nutrient genes are directly related to adaptation to environmental conditions (Figure). Variation among nutrient-related gene frequencies between sampling stations for Prochlorococcus Further, the authors found that these nutrient gene indices correlated to the measured inorganic nutrient concentrations and uptake; whereas, N gene coverage decreased with increased N uptake. Together, the inclusion of the nutrient gene indices in the trait-based models improved the predictions of carbon : phosphorus. By moving beyond strictly abiotic measurements, and using the microbiome, the authors could significantly improve predicted shifts in the elemental stoichiometry of marine communities. Papers: 1. Enquist BJ, Norberg J, Bonser SP, Violle C, Webb CT, Henderson A, Sloat LL, Savage VM. (2015). Scaling from traits to ecosystems: developing a general trait driver theory via integrating trait-based and metabolic scaling theories. Advances in Ecological Research.
DOI: 10.1016/bs.aecr.2015.02.001 2. CA Garcia, GI Hagstrom, AA Larkin, LJ Ustick, SA Levin, MW Lomas, AC Martiny. (2020). Linking regional shifts in microbial genome adaptation with surface ocean biogeochemistry. Phils Trans. R. Soc. B. DOI: 10.1098/rstb.2019.0254 3. Berube PM, Rasmussen A, Braakman R, Stepanauskas R, Chisholm SW. (2019). Emergence of trait variability through the lens of nitrogen assimilation in Prochlorococcus. eLife DOI: 10.7554/eLife.41043
0 Comments
Leave a Reply. |
AuthorSome thoughts on some (small) things Archives
May 2023
Categories |
Proudly powered by Weebly
 RSS Feed
RSS Feed