Introduction to Pangenomes
I wanted to discuss some broad discussion points with bacterial pangenomes and their genetic diversity at large. A lot of these ideas I have been discussing with the amazing microbial ecologist, Linh Anh Cat, Ph.D., and I would definitely recommend following her pieces she writes for Forbes highlighting some really interesting biological findings, including glow in the dark fungi!?
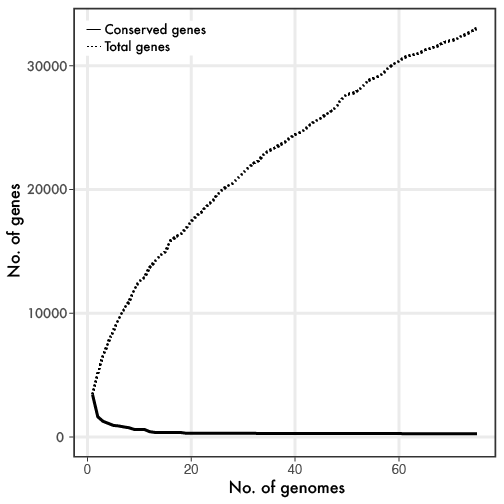
Linh Anh Cat: [Twitter link][Forbes link] With the enormous progress in sequencing microbial genomes, a general phenomenon quickly emerged revealing extensive genetic diversity in genome content, even within closely-related taxonomic groups. The utilization of the core (genes found in all members of a group) and pangenome (all genes found within a group) framework provided a baseline to examine the almost infinite genetic diversity. If you are reading this blog, you have probably seen something similar to the below figure, where the more genomes that are sampled, the smaller the core genome gets while the flexible or pangenome continually increases.
So why do prokaryotes have pangenomes?
A paper [2] a couple of years ago attempted to answer this exact question. By doing so, the authors incorporate a theoretical approach to include effective population sizes, mutation rates, selection coefficients, etc and applied neutral, deleterious, and adaptive models. They concluded that the ability of bacteria to migrate to new micro-niches enable the expansion of the pangenome. Restricted taxa (e.g., obligate intracellular organisms) have largely reduced pangenomes while free-living bacteria can have massive pangenomes. These conclusions sparked a lot of interest especially since a contemporary paper argued the complete opposite, pangenomes are directed by neutral evolution [3].
A response by BJ Shaprio [4] brought up a great point that population-level theory is tough to apply to microbial pangenomes, as there are no clear delineations of where to draw your "cut-off" (see why I thought the original tweet was so great!). Mainly, if pangenomes are driven by HGT, then transfers occur across populations, species, and broader taxonomic boundaries. One can argue for genetic relatedness and/or ecological approaches to accurately delineate where to assess pangenome composition. In the end, the paradox remained opened for debate as large effective population sizes and selection coefficients correlate both with large pangenome sizes. Why is this so interesting? For one, IF flexible genes were advantageous, we would expect selective sweeps across populations within species, thereby reducing pangenome sizes [5]. Disentangling environmental and phylogenetic constraints
So why am I bringing this all up now? Well, two recent preprints were published bringing this debate back into the forefront. To recap, the pangenome can be shaped both by habitat (via selection) and by phylogeny (via vertical inheritance). For instance, more closely-related strains will share more similar genes. However, it is really difficult to separate these two factors, phylogeny and habitat, as more closely-related strains not only share more genes, but should also prefer more similar habitats. The first of the preprint I want to highlight seeks to disentangle these effects and characterize their impact on bacterial pangenomes [6]. Using a large collection of species pangenomes (N=155 species), the authors conclude that the adaptiveness of pangenomes is partially explained by the environmental habitat and their shared core genome.
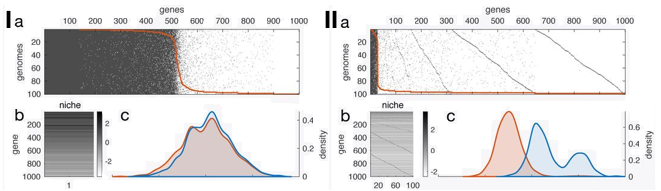
The second paper uses a more theoretical approach, a model simulation of gene content of pangenomes [7]. What I like most in this paper is that it really tried to resolve the pangenome paradox. Briefly, it comes down to the most biological conclusion, it depends. Large pangenomes of low-frequency genes are neutral. Highly beneficial genes in the pangenome can arise as a consequence of genotype-by-environment when multiple niches are available. And all if this can be influenced by the rate of gene gain and loss. In the end, we need empirical data!
I) Simulated set of 100 genomes from a single niche (1000 genes with varying fitness effects). II) Simulated set of 1 genome from 100 niches with 100 mostly-deleterious genes. A) P/A of genes in simulated pangenome B) Heatmap of sampled fitness effects C) Density plots of gene pool for theoretical (blue) and fitness effect of genes in pangenome (orange)
Papers:
1. Rocha EPC. (2018). Neutral Theory, microbial practice: Challenges in bacterial population genetics. Molecular Biology and Evolution 35(6): 1338-1347.
2. McInerney JO, McNally A, O'Connell MJ. (2017). Why prokaryotes have pangenomes. Nature Microbiology 2: 17040. 3. Andreani NA, Hesse E, Vos M. (2017). Prokaryote genome fluidity is dependent on effective population size. The ISME Journal 11: 1719-1721. 4. Shapiro BJ. (2017). The population genetics of pangenomes. Nature Microbiology 2: 1574. 5. McInerney JO, McNally A, O'Connell MJ. (2017). Reply to 'The population genetics of pangenomes'. Nature Microbiology 2:1575. 6. Maistrenko OM, Mende DR, Luetge M, Hildebrand F, Schmidt TSB, Li SS, Coelho LP, Huerta-Cepas J, Sunagawa S, Bork P. (2019). Disentangling the impact of environmental and phylogenetic constraints on prokaryotic strain diversity. bioRxiv 7. Domingo-Sananes MR, McInerney JO. (2019). Selection-based model of prokaryote pangenomes. bioRxiv
0 Comments
Leave a Reply. |
AuthorSome thoughts on some (small) things Archives
May 2023
Categories |
Proudly powered by Weebly
 RSS Feed
RSS Feed